

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MQRSNHTVTEFILLGFTTDPGMQLGLFVVFLGVYSLTVVGNSTLIVLICNDSHLHTPMYFVVGNLSFLDLWYSSVYTPKILVICISEDKSISFAGCLCQFFFSAGLAYSECCLLAAMAYDRYVAISKPLLYAQAMSIKLCALLVAVSYCGGFINSSIITKKTFSFNFCCENIIDDFFCDLLPLVKLACGEKGCYKFLMYFLLASNVICPAVLILASYLFIITSVLRISSSQGRLKAFSTCSSHLTSVTLYYGSILYIYALPRSSYSFDMDKIVSTFYTEVLPMLNPMIYSLRNKDVKEALKKLLP | |
| CCCCCCSSSSSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHCCHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCSSSSSSCCHHHCCCCHHHCCCCHHHHHHHHHHCC | |
| 99999451414676489993608999999999999999988799999862888777488887779988437111454999999714897885899999999999999999999999986514520611018823678899999999999999999999999845408899905853158288887705787266777888889999998999999999999998035675767433206588899799998544034837899888878978998811104124357440565989999986486 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MQRSNHTVTEFILLGFTTDPGMQLGLFVVFLGVYSLTVVGNSTLIVLICNDSHLHTPMYFVVGNLSFLDLWYSSVYTPKILVICISEDKSISFAGCLCQFFFSAGLAYSECCLLAAMAYDRYVAISKPLLYAQAMSIKLCALLVAVSYCGGFINSSIITKKTFSFNFCCENIIDDFFCDLLPLVKLACGEKGCYKFLMYFLLASNVICPAVLILASYLFIITSVLRISSSQGRLKAFSTCSSHLTSVTLYYGSILYIYALPRSSYSFDMDKIVSTFYTEVLPMLNPMIYSLRNKDVKEALKKLLP | |
| 85653120000000000432401000013033213313333320020010003000000100200001001000000030001002542201030000001000000100010000000100000021110103003300000011010101200201010003030144120100000320003000233301011002201321330330033033100100010312632310100110021000013201200000032431343410000210321023003000011330240033327 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCSSSSSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHCCHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCSSSSSSCCHHHCCCCHHHCCCCHHHHHHHHHHCC MQRSNHTVTEFILLGFTTDPGMQLGLFVVFLGVYSLTVVGNSTLIVLICNDSHLHTPMYFVVGNLSFLDLWYSSVYTPKILVICISEDKSISFAGCLCQFFFSAGLAYSECCLLAAMAYDRYVAISKPLLYAQAMSIKLCALLVAVSYCGGFINSSIITKKTFSFNFCCENIIDDFFCDLLPLVKLACGEKGCYKFLMYFLLASNVICPAVLILASYLFIITSVLRISSSQGRLKAFSTCSSHLTSVTLYYGSILYIYALPRSSYSFDMDKIVSTFYTEVLPMLNPMIYSLRNKDVKEALKKLLP | |||||||||||||||||||||||||
| 1 | 3emlA | 0.20 | 0.23 | 0.89 | 3.63 | Download | -------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIR | |||||||||||||||||||
| 2 | 5tgzA | 0.21 | 0.23 | 0.86 | 2.08 | Download | ------------------NPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIA------------------------VLPLLGWNCEKLQHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFSMLCLLNSTVNPIIYALRSKDLRHAFRSMF- | |||||||||||||||||||
| 3 | 5tjvA | 0.16 | 0.23 | 0.90 | 2.15 | Download | ------ENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHV-FHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLCEKLQSVCSDIF-PHID--------------------ETYLMFWIGVTSVLLLFIVYAYMYILWKAGKFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF- | |||||||||||||||||||
| 4 | 3uon | 0.15 | 0.26 | 0.90 | 1.57 | Download | -------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIV--GVRTVEDGECYIQFF---------SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINISREKKVTRTILAILLAFIITWAPYNVMVLINTFCCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM | |||||||||||||||||||
| 5 | 4yay | 0.15 | 0.23 | 0.93 | 1.28 | Download | KTTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIH-RNVFF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL- | |||||||||||||||||||
| 6 | 3emlA | 0.21 | 0.23 | 0.89 | 3.80 | Download | --------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIR | |||||||||||||||||||
| 7 | 4iaq | 0.18 | 0.24 | 0.86 | 1.72 | Download | ------------------SLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASE----------CVVNT-------D---HILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLG---IILGAFIVCWLPFFIISLVPIHL-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLI- | |||||||||||||||||||
| 8 | 4ea3A | 0.18 | 0.21 | 0.85 | 2.67 | Download | ------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLLT-LPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPTSSKAQAVNVAIWALASVVGVPVAIMGSAQVEDEIECLVEIPTPQDYWGPVFAICIFLFSF-----------------IVPVLVISVCYSLMIRRLRGVRLLSGSVAVFVGCWTPVQVFVLAQGLG-----VQPSSETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR---- | |||||||||||||||||||
| 9 | 3emlA | 0.20 | 0.23 | 0.88 | 5.27 | Download | ---------------------ISSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIR | |||||||||||||||||||
| 10 | 2ydoA | 0.16 | 0.23 | 0.92 | 6.01 | Download | ----------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIR | |||||||||||||||||||
| ||||||||||||||||||||||||||
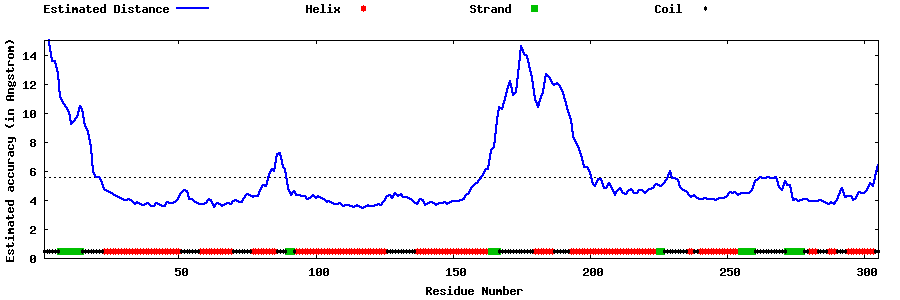
| Generated 3D models | Estimated local accuracy of models | ||
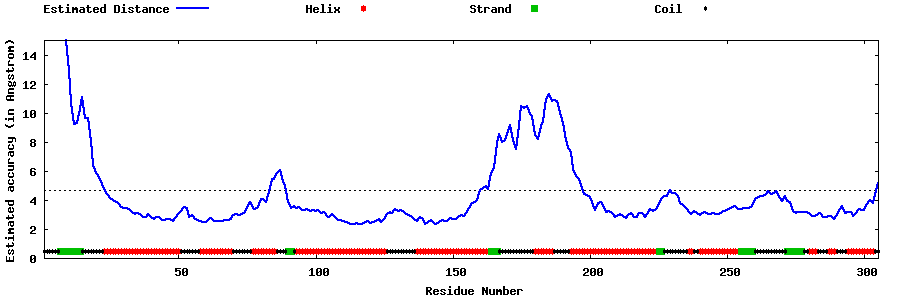 | |||
|
|
|||
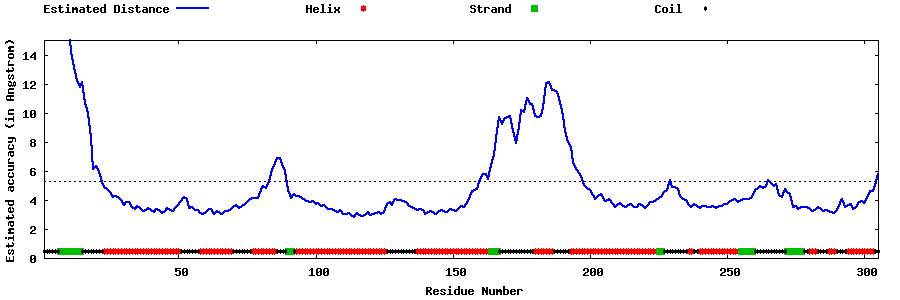 | |||
|
| |||
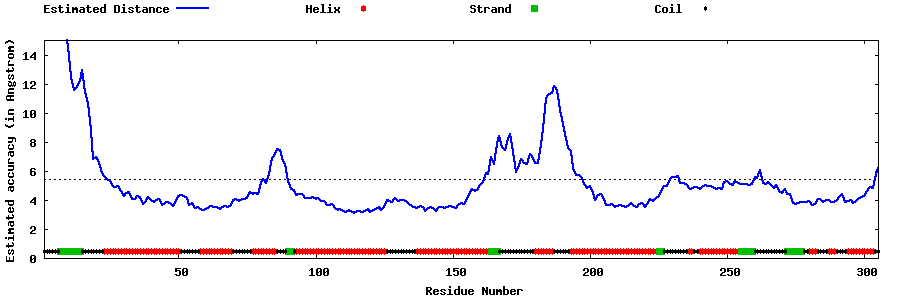 | |||
|
| |||
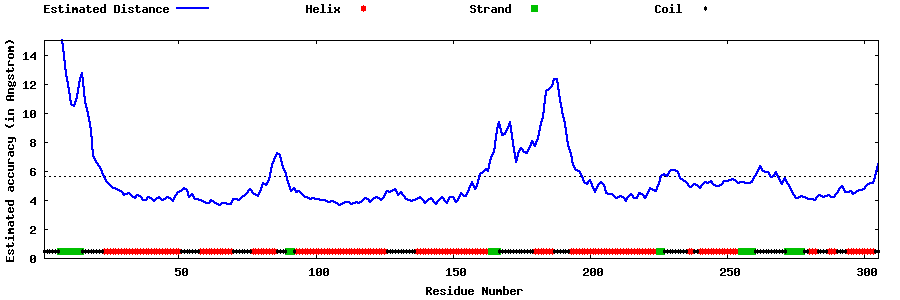 | |||
|
| |||
 | |||
|
| |||