

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MDLHLFDYSEPGNFSDISWPCNSSDCIVVDTVMCPNMPNKSVLLYTLSFIYIFIFVIGMIANSVVVWVNIQAKTTGYDTHCYILNLAIADLWVVLTIPVWVVSLVQHNQWPMGELTCKVTHLIFSINLFGSIFFLTCMSVDRYLSITYFTNTPSSRKKMVRRVVCILVWLLAFCVSLPDTYYLKTVTSASNNETYCRSFYPEHSIKEWLIGMELVSVVLGFAVPFSIIAVFYFLLARAISASSDQEKHSSRKIIFSYVVVFLVCWLPYHVAVLLDIFSILHYIPFTCRLEHALFTALHVTQCLSLVHCCVNPVLYSFINRNYRYELMKAFIFKYSAKTGLTKLIDASRVSETEYSALEQSTK | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHSSHHHHHHHHHHHHHHHHHHHSSSSSCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 99764688887777677888888877777776687763143688569999999999999889998898774366684879999999999999999729999999668989788779999999999999999999999999814766530634336464002001299999999999799998622362689847881669930356899999999999999999999999999999998647774554379999999999999967999999999999826788984799999999999999999999898999996289999999998430124786667778778788888876347789 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MDLHLFDYSEPGNFSDISWPCNSSDCIVVDTVMCPNMPNKSVLLYTLSFIYIFIFVIGMIANSVVVWVNIQAKTTGYDTHCYILNLAIADLWVVLTIPVWVVSLVQHNQWPMGELTCKVTHLIFSINLFGSIFFLTCMSVDRYLSITYFTNTPSSRKKMVRRVVCILVWLLAFCVSLPDTYYLKTVTSASNNETYCRSFYPEHSIKEWLIGMELVSVVLGFAVPFSIIAVFYFLLARAISASSDQEKHSSRKIIFSYVVVFLVCWLPYHVAVLLDIFSILHYIPFTCRLEHALFTALHVTQCLSLVHCCVNPVLYSFINRNYRYELMKAFIFKYSAKTGLTKLIDASRVSETEYSALEQSTK | |
| 74443230434321332323233232233332204334244000000021012002302321210010001133321000000000010011000000010000025330100100011001013211110000000001000000010130242232200000000011000000100000020233474322101020247214201001101211101231220002000100010242664422000000000010010013200000001002314114340422300100011010001300021000100004401420130022321353534544544434545444446648 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHSSHHHHHHHHHHHHHHHHHHHSSSSSCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MDLHLFDYSEPGNFSDISWPCNSSDCIVVDTVMCPNMPNKSVLLYTLSFIYIFIFVIGMIANSVVVWVNIQAKTTGYDTHCYILNLAIADLWVVLTIPVWVVSLVQHNQWPMGELTCKVTHLIFSINLFGSIFFLTCMSVDRYLSITYFTNTPSSRKKMVRRVVCILVWLLAFCVSLPDTYYLKTVTSASNNETYCRSFYPEHSIKEWLIGMELVSVVLGFAVPFSIIAVFYFLLARAISASSDQEKHSSRKIIFSYVVVFLVCWLPYHVAVLLDIFSILHYIPFTCRLEHALFTALHVTQCLSLVHCCVNPVLYSFINRNYRYELMKAFIFKYSAKTGLTKLIDASRVSETEYSALEQSTK | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.27 | 0.22 | 0.81 | 3.27 | Download | --------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKE--GLHYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.27 | 0.22 | 0.81 | 4.27 | Download | --------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKE--GLHYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFF-------------------------------- | |||||||||||||||||||
| 3 | 4mbsA | 0.27 | 0.22 | 0.81 | 3.91 | Download | --------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEG--LHYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.27 | 0.23 | 0.77 | 1.56 | Download | ---------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------------ | |||||||||||||||||||
| 5 | 4djh | 0.27 | 0.23 | 0.77 | 1.18 | Download | ---------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS------A------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------------ | |||||||||||||||||||
| 6 | 4mbsA | 0.27 | 0.22 | 0.81 | 3.31 | Download | ---------------------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQ--WDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEG--LHYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------- | |||||||||||||||||||
| 7 | 4djh | 0.27 | 0.23 | 0.77 | 1.73 | Download | -----------------------------------------AIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVY-LMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.27 | 0.22 | 0.81 | 4.64 | Download | --------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQ--WDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGL--HYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRE--KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.28 | 0.22 | 0.81 | 3.60 | Download | --------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKE--GLHYTCSSHFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEKHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.25 | 0.22 | 0.79 | 4.18 | Download | ----------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMG-TWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQ--GSIDCTLTFSHP-TWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPET-------TFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF------------------------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
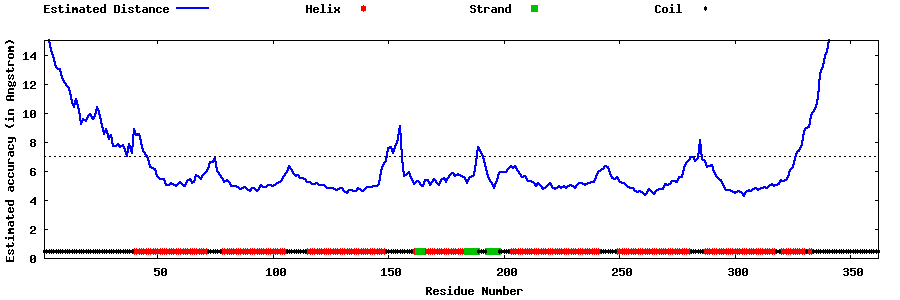
| Generated 3D models | Estimated local accuracy of models | ||
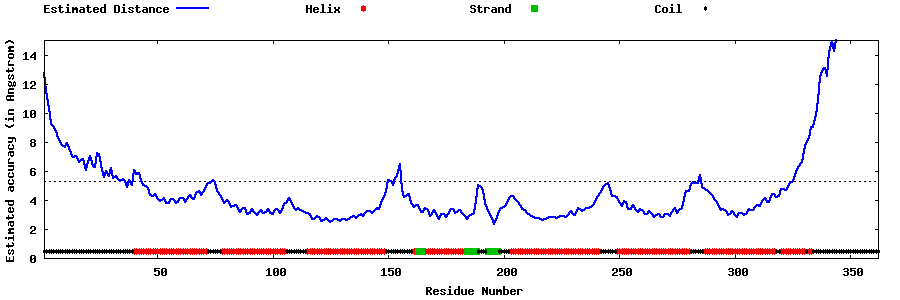 | |||
|
|
|||
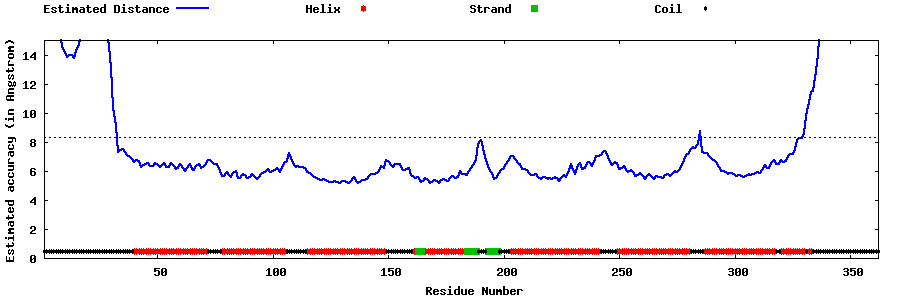 | |||
|
| |||
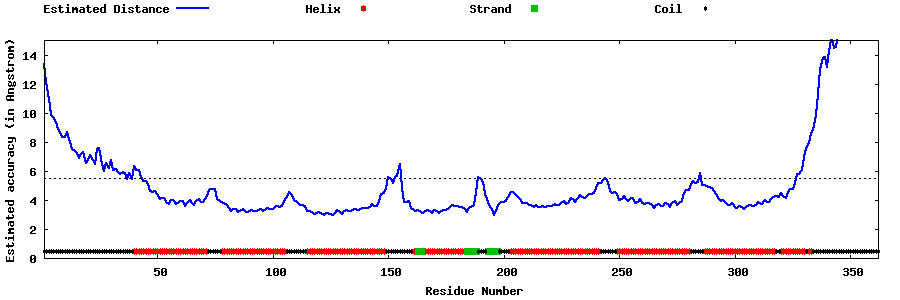 | |||
|
| |||
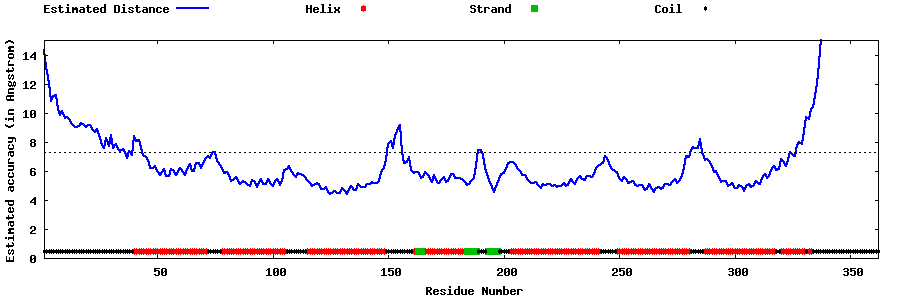 | |||
|
| |||
 | |||
|
| |||