

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MSPLNQSAEGLPQEASNRSLNATETSEAWDPRTLQALKISLAVVLSVITLATVLSNAFVLTTILLTRKLHTPANYLIGSLATTDLLVSILVMPISIAYTITHTWNFGQILCDIWLSSDITCCTASILHLCVIALDRYWAITDALEYSKRRTAGHAATMIAIVWAISICISIPPLFWRQAKAQEEMSDCLVNTSQISYTIYSTCGAFYIPSVLLIILYGRIYRAARNRILNPPSLYGKRFTTAHLITGSAGSSLCSLNSSLHEGHSHSAGSPLFFNHVKIKLADSALERKRISAARERKATKILGIILGAFIICWLPFFVVSLVLPICRDSCWIHPALFDFFTWLGYLNSLINPIIYTVFNEEFRQAFQKIVPFRKAS | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCSSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCSSSSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHCSSHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHCCCCCCC | |
| 99876677778777778777777778888887646899999999999999999999871564560167775767999999999999999987799999982506787489999999999999999999999999748855644018763149999999999999999999999981876678889807998457159999999999999999999999999999998860232334432233334444334554332222223444434432345543235665433110333577767651000014589997899999999998088751788999999999999983868999837899999999982877899 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MSPLNQSAEGLPQEASNRSLNATETSEAWDPRTLQALKISLAVVLSVITLATVLSNAFVLTTILLTRKLHTPANYLIGSLATTDLLVSILVMPISIAYTITHTWNFGQILCDIWLSSDITCCTASILHLCVIALDRYWAITDALEYSKRRTAGHAATMIAIVWAISICISIPPLFWRQAKAQEEMSDCLVNTSQISYTIYSTCGAFYIPSVLLIILYGRIYRAARNRILNPPSLYGKRFTTAHLITGSAGSSLCSLNSSLHEGHSHSAGSPLFFNHVKIKLADSALERKRISAARERKATKILGIILGAFIICWLPFFVVSLVLPICRDSCWIHPALFDFFTWLGYLNSLINPIIYTVFNEEFRQAFQKIVPFRKAS | |
| 74332322432335233332231334443444222011000001113103203311200000001134022100000000010011000000230000001230211310001000000300000020000001000000020030434223100000000001100100000000021446643430102244210000002100310000002001200200242244245333222232323323343433432333322223322223332332232232222332232234331132100000001201211021000200034302013100000012012000000000001043014002300203268 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCSSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCSSSSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHCSSHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHCCCCCCC MSPLNQSAEGLPQEASNRSLNATETSEAWDPRTLQALKISLAVVLSVITLATVLSNAFVLTTILLTRKLHTPANYLIGSLATTDLLVSILVMPISIAYTITHTWNFGQILCDIWLSSDITCCTASILHLCVIALDRYWAITDALEYSKRRTAGHAATMIAIVWAISICISIPPLFWRQAKAQEEMSDCLVNTSQISYTIYSTCGAFYIPSVLLIILYGRIYRAARNRILNPPSLYGKRFTTAHLITGSAGSSLCSLNSSLHEGHSHSAGSPLFFNHVKIKLADSALERKRISAARERKATKILGIILGAFIICWLPFFVVSLVLPICRDSCWIHPALFDFFTWLGYLNSLINPIIYTVFNEEFRQAFQKIVPFRKAS | |||||||||||||||||||||||||
| 1 | 4iaqA | 0.57 | 0.53 | 0.89 | 2.47 | Download | --------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFWRQASE------CVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADLEDNWETLNDNLKVIAQKATPDSPEMKDFRHGFDIGKVKEAQAAAEQLKTTRNAYIQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIH-------LAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| 2 | 4ib4A | 0.26 | 0.28 | 0.90 | 4.61 | Download | -------------------------------------LHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIETNPNNITCVLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKKQAAAEQLKTTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR- | |||||||||||||||||||
| 3 | 5uenA | 0.22 | 0.24 | 0.91 | 3.34 | Download | --------------------------------SISAFQAAYIGIEVLIALVSVPGNVLVIWAVKVNQALRDATFCFIVSLAVADVAVGALVIPLAILINIGPQTYF--HTCLMVACPVLILTQSSILALLAIAVDRYLRVKIPLRYKMVVTPRRAAVAIAGCWILSFVVGLTPMFGWNNAGSMGEPVIKCEISMEYMVYFNFFVWVLPPLLLMVLIYLEVFYLIRKQLADLEDNNVKDALTKMRAPEMKDFRHGFDILVGEGKVKEAQAAAEQLKTTRNAYIQKYLERARSTLQKELKIAKSLALILFLFALSWLPLHILNCITLFCPSC-HKPSILTYIAIFLTHGNSAMNPIVYAFRIQKFRVTFLKIWNDHFRC | |||||||||||||||||||
| 4 | 4ib4 | 0.26 | 0.28 | 0.91 | 1.55 | Download | -------------------------EEQGN------KLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIET-NPNNITCVLTKERGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLKVIEKADNAQLTRAAALDQKGFDINEGKVEAQAAAQLKTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR- | |||||||||||||||||||
| 5 | 3uon | 0.29 | 0.29 | 0.89 | 1.23 | Download | ----------------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVEDGECYIQFSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMLRIDEGLRLKIYKPSAIGRNTNGVITKDRGILRNAKQQKRDEANLAKSFRTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAP-C-IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM----- | |||||||||||||||||||
| 6 | 4ib4A | 0.25 | 0.28 | 0.91 | 3.16 | Download | --------------------------------EQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIETNPNNITCVLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQEAQAAAEQLKTTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR- | |||||||||||||||||||
| 7 | 4ib4 | 0.26 | 0.28 | 0.89 | 1.74 | Download | --------------------------------------HWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIGIETN--PNNITCVLTKERGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLKVIEKADNAAQVKMRAAALKGFDINEGKVKEAQAEQLKTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR- | |||||||||||||||||||
| 8 | 4iarA | 0.55 | 0.55 | 0.89 | 4.75 | Download | --------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFWRQASECVVNT------DHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMYIQKYLAARERKATKTLGIILGAFIVCWLPFFIISLVMPIWFHL-----AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| 9 | 4iaqA | 0.57 | 0.53 | 0.89 | 2.86 | Download | --------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFWRQA------SECVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADLEDNWETLNDNLKVINAAQVKDALTKMRAAALDAQKATPDSPEAAEQLKTTRNAYIQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIH-------LAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| 10 | 4iaqA | 0.57 | 0.53 | 0.89 | 2.80 | Download | --------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFWRQ------ASECVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADLEDNWETLNDNLKVINAAQVKDALTKMRAAALDAQKATPDSPEMKDFRHTTRNAYIQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLV-------MPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| ||||||||||||||||||||||||||
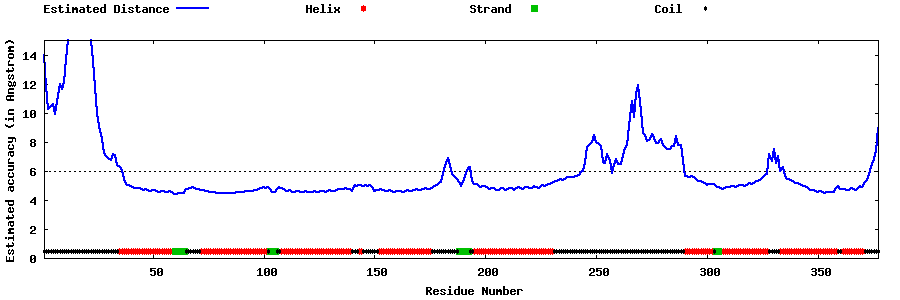
| Generated 3D models | Estimated local accuracy of models | ||
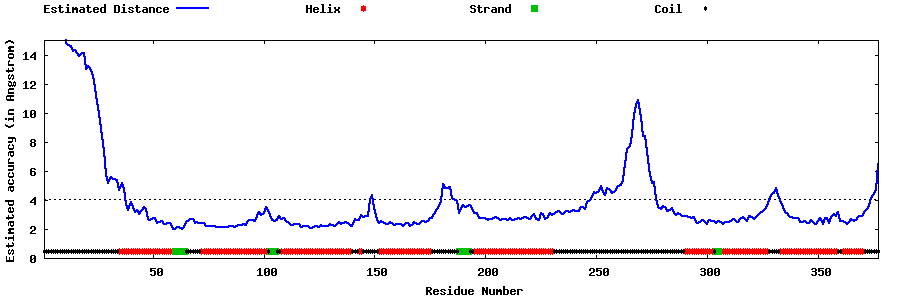 | |||
|
|
|||
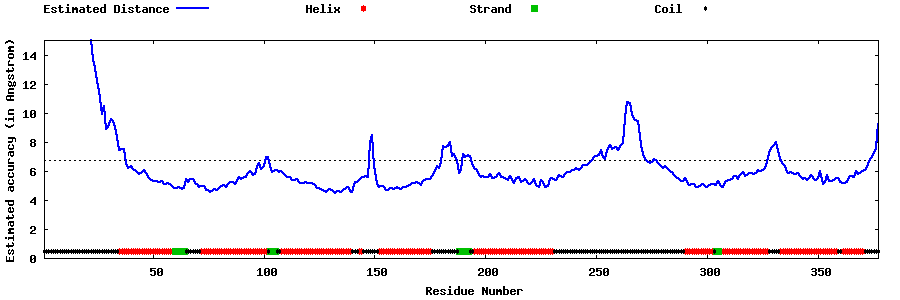 | |||
|
| |||
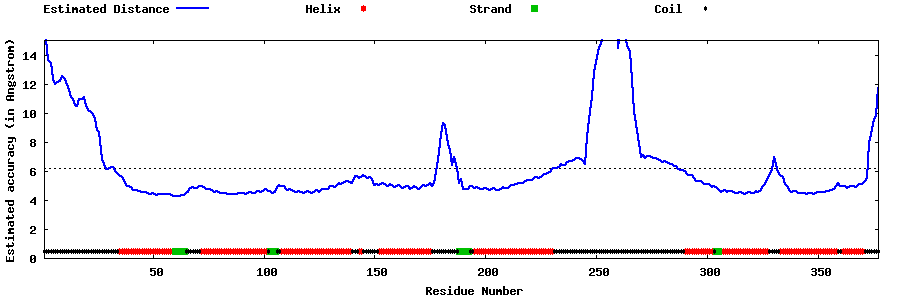 | |||
|
| |||
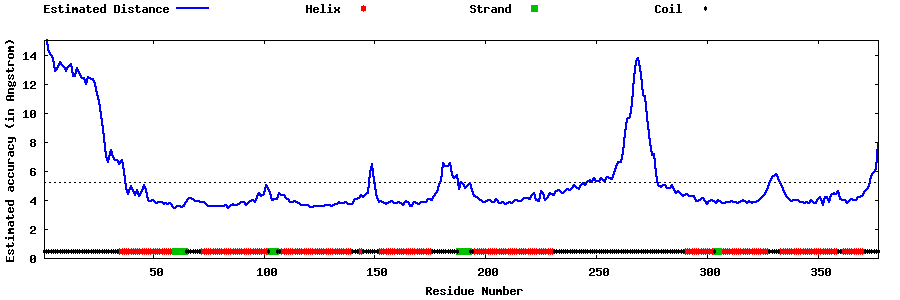 | |||
|
| |||
 | |||
|
| |||