

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MNITNCTTEASMAIRPKTITEKMLICMTLVVITTLTTLLNLAVIMAIGTTKKLHQPANYLICSLAVTDLLVAVLVMPLSIIYIVMDRWKLGYFLCEVWLSVDMTCCTCSILHLCVIALDRYWAITNAIEYARKRTAKRAALMILTVWTISIFISMPPLFWRSHRRLSPPPSQCTIQHDHVIYTIYSTLGAFYIPLTLILILYYRIYHAAKSLYQKRGSSRHLSNRSTDSQNSFASCKLTQTFCVSDFSTSDPTTEFEKFHASIRIPPFDNDLDHPGERQQISSTRERKAARILGLILGAFILSWLPFFIKELIVGLSIYTVSSEVADFLTWLGYVNSLINPLLYTSFNEDFKLAFKKLIRCREHT | |
| CCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCSSSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHCCCCCCC | |
| 99988899888788998889999999999999999999898732225601566754589999999999999999866999999838067887899999999999999999999999986551785511278643489999989999999999999999838866666788825898617178869999999999999999999999999998654203333334433222334332222232244543233322223334332223345542212202333446788999999999999876602899999998744788976999999999999983759999808999999999972886899 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MNITNCTTEASMAIRPKTITEKMLICMTLVVITTLTTLLNLAVIMAIGTTKKLHQPANYLICSLAVTDLLVAVLVMPLSIIYIVMDRWKLGYFLCEVWLSVDMTCCTCSILHLCVIALDRYWAITNAIEYARKRTAKRAALMILTVWTISIFISMPPLFWRSHRRLSPPPSQCTIQHDHVIYTIYSTLGAFYIPLTLILILYYRIYHAAKSLYQKRGSSRHLSNRSTDSQNSFASCKLTQTFCVSDFSTSDPTTEFEKFHASIRIPPFDNDLDHPGERQQISSTRERKAARILGLILGAFILSWLPFFIKELIVGLSIYTVSSEVADFLTWLGYVNSLINPLLYTSFNEDFKLAFKKLIRCREHT | |
| 61412322435343442300110010121132033033112000000011330231000000000200120010002310000012302103100010000001000000200000020001000200303342231000000000021011001000000224544344430203234300000002101310000002001100100122134344333333344433233322322232322333333243333333332333333333233323332323443300200000101110012210000001000532014100000012013001100000020043014002300203338 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCSSSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHCCCCCCC MNITNCTTEASMAIRPKTITEKMLICMTLVVITTLTTLLNLAVIMAIGTTKKLHQPANYLICSLAVTDLLVAVLVMPLSIIYIVMDRWKLGYFLCEVWLSVDMTCCTCSILHLCVIALDRYWAITNAIEYARKRTAKRAALMILTVWTISIFISMPPLFWRSHRRLSPPPSQCTIQHDHVIYTIYSTLGAFYIPLTLILILYYRIYHAAKSLYQKRGSSRHLSNRSTDSQNSFASCKLTQTFCVSDFSTSDPTTEFEKFHASIRIPPFDNDLDHPGERQQISSTRERKAARILGLILGAFILSWLPFFIKELIVGLSIYTVSSEVADFLTWLGYVNSLINPLLYTSFNEDFKLAFKKLIRCREHT | |||||||||||||||||||||||||
| 1 | 4iaqA | 0.48 | 0.48 | 0.93 | 2.65 | Download | ----------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFWR-------QASECVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADLEDNWETLNDNLKVINAAQVKDALTKMRAAALDAQKATPDSPEMKAQAAAEQLKTTRNAYIQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMP-----IHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| 2 | 5uenA | 0.22 | 0.26 | 0.94 | 4.61 | Download | -------------------AFQAAYIGIEVLIALVSVPGNVLVIWAVKVNQALRDATFCFIVSLAVADVAVGALVIPLAILINIGPQTYFH--TCLMVACPVLILTQSSILALLAIAVDRYLRVKIPLRYKMVVTPRRAAVAIAGCWILSFVVGLTPMFGWNNLSAVERKCEFEKVISMEYMVYFNFFVWVLPPLLLMVLIYLEVFYLIRKQLADLEDNWETLNDNVKDALTKMRAAALDAPEMKDFRANEGKVKEAQAAAEQLKTTRNAYIQKYLERARSTLQKELKIAKSLALILFLFALSWLPLHILNCITLFCPSCKPSILTYIAIFLTHGNSAMNPIVYAFRIQKFRVTFLKIWNDHFR- | |||||||||||||||||||
| 3 | 5uenA | 0.21 | 0.26 | 0.95 | 3.56 | Download | ----------------SISAFQAAYIGIEVLIALVSVPGNVLVIWAVKVNQALRDATFCFIVSLAVADVAVGALVIPLAILINIGPQTYF--HTCLMVACPVLILTQSSILALLAIAVDRYLRVKIPLRYKMVVTPRRAAVAIAGCWILSFVVGLTPMFGWNNLSAAGSMGEPVIKCEMEYMVYFNFFVWVLPPLLLMVLIYLEVFYLIRKQLADLEDNWETLNDN-VKDALTKMRAAALDAPEMKDFRHGFDILVGQIDDALKLATRNAYIQKYLERARSTLQKELKIAKSLALILFLFALSWLPLHILNCITLFCPSCKPSILTYIAIFLTHGNSAMNPIVYAFRIQKFRVTFLKIWNDHFRC | |||||||||||||||||||
| 4 | 4ib4 | 0.27 | 0.30 | 0.95 | 1.56 | Download | ----------EEQ-----GNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIETN--PNNITCVLTKERGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNEKADNAAQMRAADAQKKDRGFDIVGQIDLANEGKVKEAQAAAKTTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR- | |||||||||||||||||||
| 5 | 3uon | 0.25 | 0.28 | 0.94 | 1.25 | Download | ------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVTVEDGECYIQSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMLRIDEGLRLKEGKSPSLNAAIGRNTNGVITKINAKDSLAGLQQKLAKSRVITTFRTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM----- | |||||||||||||||||||
| 6 | 4iaqA | 0.47 | 0.48 | 0.93 | 3.34 | Download | -----------IYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFWR-------QASECVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADLEDNWETLNDNLKVINAAQVKDALTKMRAAALDAQKATPDSPEMKDFRHGFDIKTTRNAYIQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPI-----HLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| 7 | 4ib4 | 0.27 | 0.30 | 0.93 | 1.77 | Download | ---------------------LHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPPIKGIETN---PNNITCVLTKERGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLKVIEKADAAQVMRAAALDAQKKDRHGFDANEGKVKEAQAAEQTTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR- | |||||||||||||||||||
| 8 | 4iarA | 0.46 | 0.47 | 0.94 | 4.64 | Download | ----------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFWRQASECVVNTD-------HILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDRNAYIQKYLAARERKATKTLGIILGAFIVCWLPFFIISLVM---PIWFHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| 9 | 4iaqA | 0.47 | 0.48 | 0.93 | 3.03 | Download | ----------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFWRQASE-------CVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADLEDNWETLNDNLKVINAAQVKDALTKMRAAALDAQKATPDSPEMKAQAAAEQLKTTRNAYIQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMP-----IHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| 10 | 5cxvA | 0.27 | 0.96 | 3.65 | Download | ----------------KGPWQVAFIGITTGLLSLATVTGNLLVLISFKVNTELKTVNNYFLLSLACADLIIGTFSMNLYTTYLLMGHWALGTLACDLWLALDYVASQASVMNLLLISFDRYFSVTRPLSYRAKRTPRRAALMIGLAWLVSFVLWAPAILFWQYERTVLAGQCYIQFLSQPIITFGTAMAAFYLPVTVMCTLYWRIYRETENRNIFEMLRIDEGLRLKIYKDTEGYYTIGIGHLLTKSPSLNAAKSELDKAIGRNTNGVITKFRTGTWDAYFSLVKEKKAARTLSAILLAFILTWTPYNIMVLVSTFCKDCVPETLWELGYWLCYVNSTINPMCYALCNKAFRDTFRLLLLCRWDK | ||||||||||||||||||||
| ||||||||||||||||||||||||||
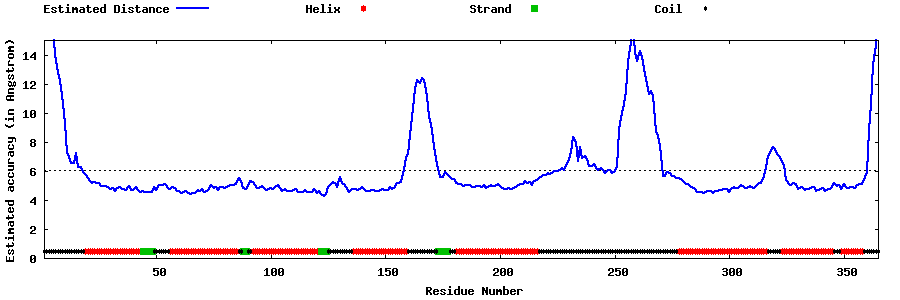
| Generated 3D models | Estimated local accuracy of models | ||
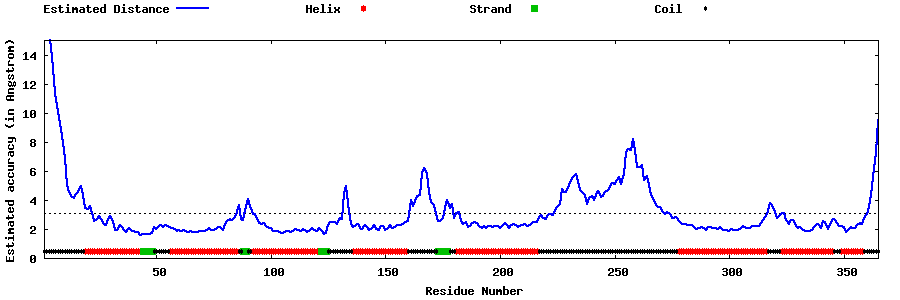 | |||
|
|
|||
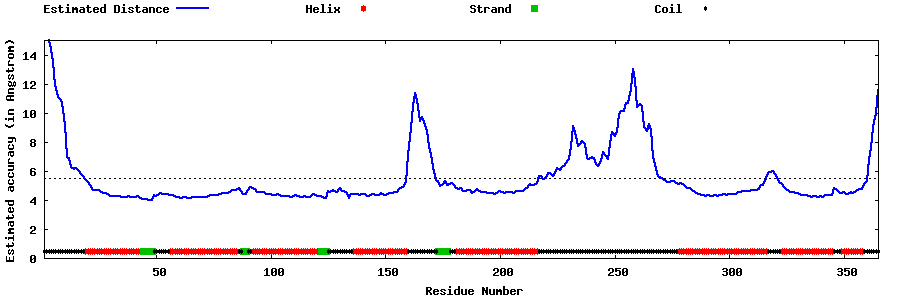 | |||
|
| |||
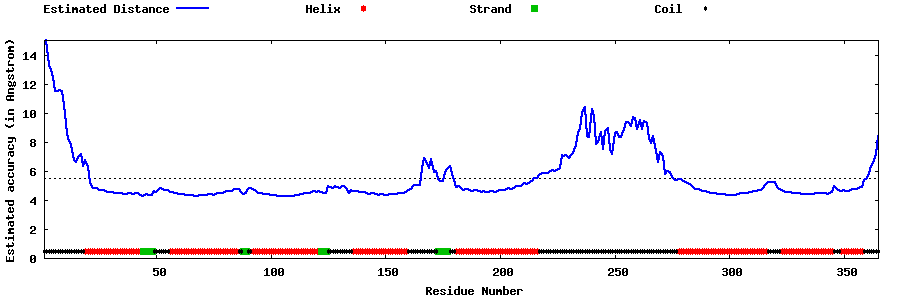 | |||
|
| |||
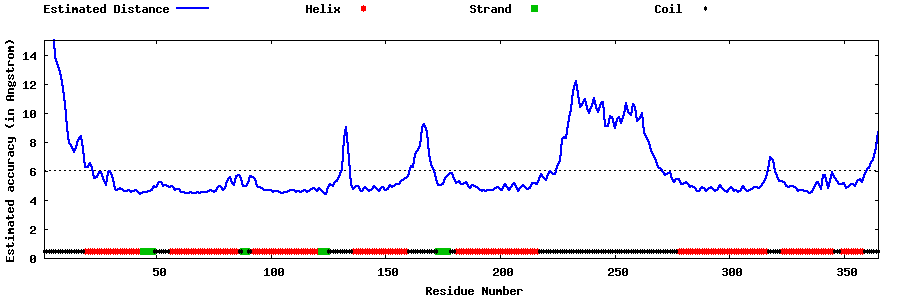 | |||
|
| |||
 | |||
|
| |||