

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MLLETQDALYVALELVIAALSVAGNVLVCAAVGTANTLQTPTNYFLVSLAAADVAVGLFAIPFAITISLGFCTDFYGCLFLACFVLVLTQSSIFSLLAVAVDRYLAICVPLRYKSLVTGTRARGVIAVLWVLAFGIGLTPFLGWNSKDSATNNCTEPWDGTTNESCCLVKCLFENVVPMSYMVYFNFFGCVLPPLLIMLVIYIKIFLVACRQLQRTELMDHSRTTLQREIHAAKSLAMIVGIFALCWLPVHAVNCVTLFQPAQGKNKPKWAMNMAILLSHANSVVNPIVYAYRNRDFRYTFHKIISRYLLCQADVKSGNGQAGVQPALGVGL | |
| CCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHSSHHHHHHHHHHHHHHHHHHSSSCCCCCCCSSCCCCCCCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCC | |
| 98769999999999999999999899753205716988858999999999999999999999999999632687670809999999999999999999999738451263779742838867260319999999999999999420003665412136787775237751352024666899999999999999999999999999999773477766542067888653799999999999999979999999999957643577229999999999999999999999957999999999997164278998998888899999999999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MLLETQDALYVALELVIAALSVAGNVLVCAAVGTANTLQTPTNYFLVSLAAADVAVGLFAIPFAITISLGFCTDFYGCLFLACFVLVLTQSSIFSLLAVAVDRYLAICVPLRYKSLVTGTRARGVIAVLWVLAFGIGLTPFLGWNSKDSATNNCTEPWDGTTNESCCLVKCLFENVVPMSYMVYFNFFGCVLPPLLIMLVIYIKIFLVACRQLQRTELMDHSRTTLQREIHAAKSLAMIVGIFALCWLPVHAVNCVTLFQPAQGKNKPKWAMNMAILLSHANSVVNPIVYAYRNRDFRYTFHKIISRYLLCQADVKSGNGQAGVQPALGVGL | |
| 75242211010201220122023332000000010340100000002010302312000101100001031021111001100000000000130300001300100010021333123310000000011100200110000122343334323331433344310102121322233211120223331331010002002200110132044354464444333342100000000020112111200000000002322333002000100121012100021000000044023001100012002344444444444443345356 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHSSHHHHHHHHHHHHHHHHHHSSSCCCCCCCSSCCCCCCCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCC MLLETQDALYVALELVIAALSVAGNVLVCAAVGTANTLQTPTNYFLVSLAAADVAVGLFAIPFAITISLGFCTDFYGCLFLACFVLVLTQSSIFSLLAVAVDRYLAICVPLRYKSLVTGTRARGVIAVLWVLAFGIGLTPFLGWNSKDSATNNCTEPWDGTTNESCCLVKCLFENVVPMSYMVYFNFFGCVLPPLLIMLVIYIKIFLVACRQLQRTELMDHSRTTLQREIHAAKSLAMIVGIFALCWLPVHAVNCVTLFQPAQGKNKPKWAMNMAILLSHANSVVNPIVYAYRNRDFRYTFHKIISRYLLCQADVKSGNGQAGVQPALGVGL | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.22 | 0.26 | 0.88 | 2.61 | Download | ASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMTWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD------------GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV----RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKP------------------------ | |||||||||||||||||||
| 2 | 4amjA | 0.34 | 0.32 | 0.86 | 4.39 | Download | ----QWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWR-----------DEDPQALKCYQDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQIRKIDRRKTSRVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNR---DLVPDWLFVAFNWLGYANSAMNPIIYCR-SPDFRKAFKRLL--------------------------- | |||||||||||||||||||
| 3 | 3sn6R | 0.32 | 0.32 | 0.85 | 3.79 | Download | ---EVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQM----HWYRQEAINCYAEET-------CCDF---FTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGRC---LKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQ---DNLIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLC-------------------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.26 | 0.28 | 0.88 | 1.57 | Download | EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPKGIETN-PNNITCVLTK-----------------ERFGDFML-FGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR---------------------- | |||||||||||||||||||
| 5 | 4djh | 0.22 | 0.24 | 0.87 | 1.16 | Download | --SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVT-TTMPFQSTVYLSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRED----------VDVIECSLQFPDDDSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNITAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.23 | 0.26 | 0.88 | 3.09 | Download | --SLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD------------GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV----RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------- | |||||||||||||||||||
| 7 | 3uon | 0.24 | 0.25 | 0.86 | 1.73 | Download | ----FEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTV---------EDGECYIQFFSN-----AAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEGAADAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAP---CIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM-------------------------- | |||||||||||||||||||
| 8 | 3emlA | 0.62 | 0.57 | 0.88 | 4.75 | Download | MGS----SVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS----TGFHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGL-----------TPMLGWNNCGQSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLNRTGTWDAYRSTLQKEVHAAKSLAIIVGLFALCWLPLHIINCFTFFCPDCSHA-PLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-------------------- | |||||||||||||||||||
| 9 | 3sn6R | 0.32 | 0.32 | 0.85 | 3.12 | Download | ---EVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRQEAIN--------------CYAEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGR---CLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQD---NLIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLC-------------------------- | |||||||||||||||||||
| 10 | 4ug2A | 0.61 | 0.56 | 0.88 | 4.48 | Download | ------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIAISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVA----CLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQL--------ARSTLQKEVHAAKSLAIIVGLFALCWLPLHIINCFTFFCPDC-SHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR--------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
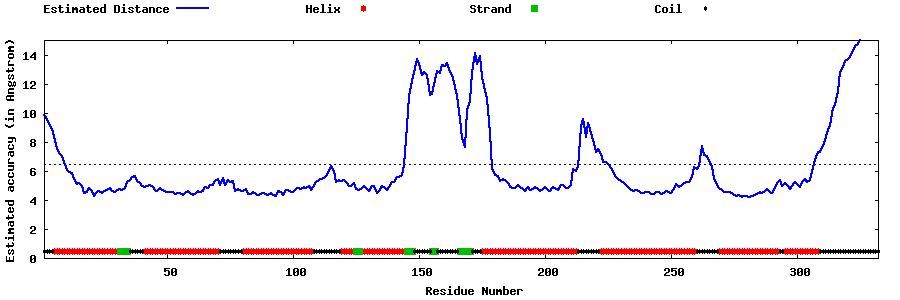
| Generated 3D models | Estimated local accuracy of models | ||
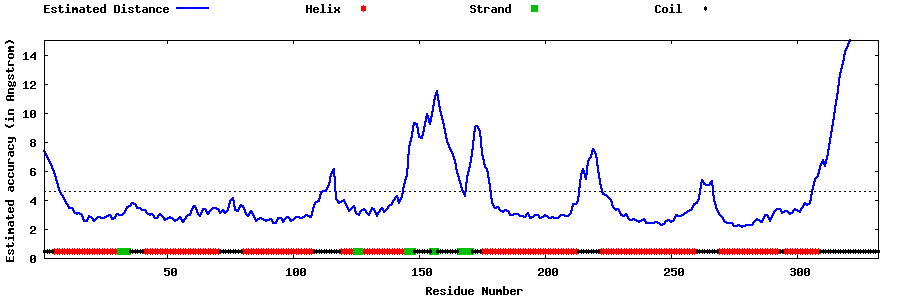 | |||
|
|
|||
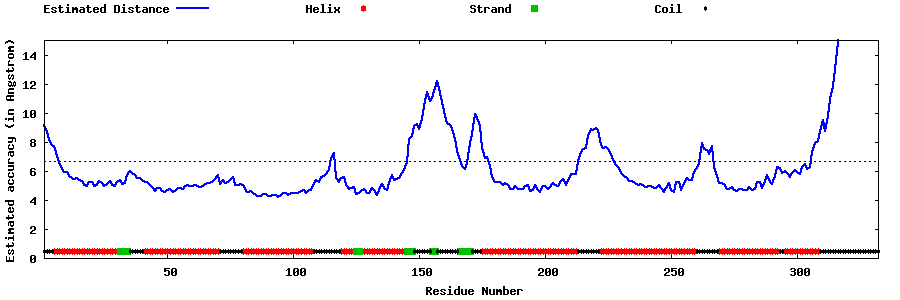 | |||
|
| |||
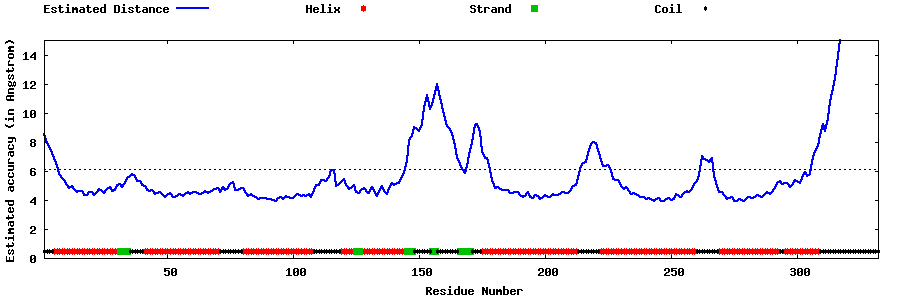 | |||
|
| |||
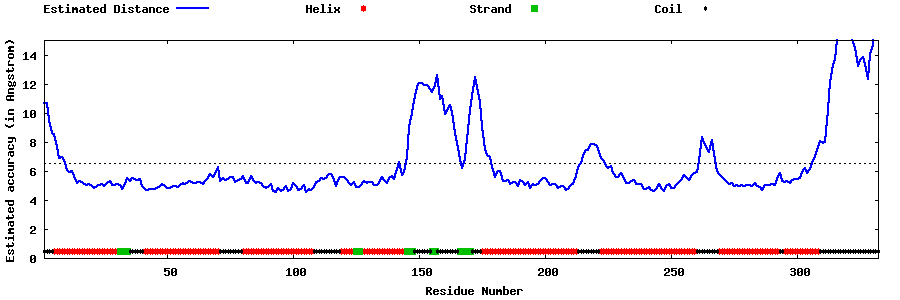 | |||
|
| |||
 | |||
|
| |||