

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MSMNNSKQLVSPAAALLSNTTCQTENRLSVFFSVIFMTVGILSNSLAIAILMKAYQRFRQKSKASFLLLASGLVITDFFGHLINGAIAVFVYASDKEWIRFDQSNVLCSIFGICMVFSGLCPLLLGSVMAIERCIGVTKPIFHSTKITSKHVKMMLSGVCLFAVFIALLPILGHRDYKIQASRTWCFYNTEDIKDWEDRFYLLLFSFLGLLALGVSLLCNAITGITLLRVKFKSQQHRQGRSHHLEMVIQLLAIMCVSCICWSPFLVTMANIGINGNHSLETCETTLFALRMATWNQILDPWVYILLRKAVLKNLYKLASQCCGVHVISLHIWELSSIKNSLKVAAISESPVAEKSAST | |
| CCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSSSCCCSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 96888898899987677888643110699999999999999999999997785662058998722899999999999999999889999999727940254897463455999999999999999999999989992063523568356355655779999999999887668836999789449947289975267889788999999999999999999999999997711355410007899999999899999999989999999999976877767305999999999999998599999349999999999988755888767667888775788735666899998899999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MSMNNSKQLVSPAAALLSNTTCQTENRLSVFFSVIFMTVGILSNSLAIAILMKAYQRFRQKSKASFLLLASGLVITDFFGHLINGAIAVFVYASDKEWIRFDQSNVLCSIFGICMVFSGLCPLLLGSVMAIERCIGVTKPIFHSTKITSKHVKMMLSGVCLFAVFIALLPILGHRDYKIQASRTWCFYNTEDIKDWEDRFYLLLFSFLGLLALGVSLLCNAITGITLLRVKFKSQQHRQGRSHHLEMVIQLLAIMCVSCICWSPFLVTMANIGINGNHSLETCETTLFALRMATWNQILDPWVYILLRKAVLKNLYKLASQCCGVHVISLHIWELSSIKNSLKVAAISESPVAEKSAST | |
| 46163444333334322332223322100011123213313322330110000023436344301000000000200020023113200000023230221220220002001010211210120000002000000000122343033310000000001200200210100002122233100000103345312100000101311321021013012100100131244345454434311200000000010011013101200000002333332212010100200020003002000001430041014002300324335463344433444444443454446564667 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSSSCCCSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MSMNNSKQLVSPAAALLSNTTCQTENRLSVFFSVIFMTVGILSNSLAIAILMKAYQRFRQKSKASFLLLASGLVITDFFGHLINGAIAVFVYASDKEWIRFDQSNVLCSIFGICMVFSGLCPLLLGSVMAIERCIGVTKPIFHSTKITSKHVKMMLSGVCLFAVFIALLPILGHRDYKIQASRTWCFYNTEDIKDWEDRFYLLLFSFLGLLALGVSLLCNAITGITLLRVKFKSQQHRQGRSHHLEMVIQLLAIMCVSCICWSPFLVTMANIGINGNHSLETCETTLFALRMATWNQILDPWVYILLRKAVLKNLYKLASQCCGVHVISLHIWELSSIKNSLKVAAISESPVAEKSAST | |||||||||||||||||||||||||
| 1 | 4zwjA | 0.15 | 0.15 | 0.97 | 2.52 | Download | PFSNATGVVRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQH-KKLRTPLN----YILLNLAVADLFMVLGGFTSTLYTSLHG----YFVFGPTGCNLQGFFATLGGEIALWSLVVLAIERYVVVCKPMSN-FRFGENHAIMGVAFTWVMALACAAPPLAGWSRYIPEGLQCSCGIDYYTLEVNNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIYVIAFLICWVPYASVAFYIFTHQGSCF-GPIFMTIPAFFAKSAAIYNPVIYIMMNKQFRNCMLTTICCGKIFKKVSRDKSVTIYLGKRDYVDHVSQVEPDPELVKG | |||||||||||||||||||
| 2 | 4bwbA | 0.13 | 0.16 | 0.82 | 3.91 | Download | -------------NSDLDVNTDIYSKVLVTAIYLALFVVGTVGNGVTLFTL-------AQSLQSRVDYYLGSLALSDLLILLFALPVDLYNFIWVHH--PWAFGDAGCKGYYFLREACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGLVCT--PIVDTATLRVVIQLNTFMSFLFPMLVASILNTVAARRLTVM-----VEPGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFVYISDEQWTTHYFYMLSNALVYVSAAINPILYNLVSANFRQVFLSTLACLC------------------------------------ | |||||||||||||||||||
| 3 | 2z73A | 0.14 | 0.18 | 0.94 | 3.53 | Download | ETWWYNPSIVVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKT-KSLQTP----ANMFIINLAFSDFTFSLVNGPLMTISCFLK----KWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWV-TPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLT-CCQFDDKETEDDKDAETEIPAGE----------- | |||||||||||||||||||
| 4 | 2ziy | 0.16 | 0.20 | 0.97 | 1.55 | Download | DLRDNETWWYNPSIIVEFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTK-TKSLQ----TPANMFIINLAFSDFTFSLVNFPLMTISC-FLKKWI---FGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLE-WVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETEDDKDAETEIP--AGESSDAAPSADA | |||||||||||||||||||
| 5 | 2ziy | 0.13 | 0.20 | 0.97 | 1.17 | Download | -DLRDNETWWYNPSIREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQ-----TPANMFIINLAFSDFTFSLVGFPLMTISCFLK----KWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPL-EWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVCCQFDDKETEDDKDAETEIPAGESSDAAPSADAAQMK | |||||||||||||||||||
| 6 | 2ziyA | 0.14 | 0.20 | 0.97 | 2.91 | Download | ETWWYNPSIIVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKT-KSLQTP----ANMFIINLAFSDFTFSLVNGPLMTISCFLK----KWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWV-TPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETEDDKDAETEIPAGESSDAAPSADAAQ | |||||||||||||||||||
| 7 | 3uon | 0.13 | 0.18 | 0.77 | 1.68 | Download | ---------------------------FIVLVAGSLSLVTIIGNILVMVSIKVN---RHLQ--TVNNYFLFSLACADLIIGVFSMNLYTLYTV-IGY---WPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVVEDGECYIQFFS----NAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIEGDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPC--IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLL---------------------------------------- | |||||||||||||||||||
| 8 | 1u19A | 0.16 | 0.19 | 0.93 | 4.58 | Download | FS-NKTGVVRSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQH-----KKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHG----YFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPM-SNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPHENNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFG-PIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLC--CGKNPLGDD--EASTTVSKTETSQVAPA--------- | |||||||||||||||||||
| 9 | 2ziyA | 0.13 | 0.20 | 0.97 | 2.55 | Download | LRDNETPSIIVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKT-KSLQTP----ANMFIINLAFSDFTFSLVNFPLMTISCFLK----KWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPL-EWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETEDDKDAETEIPAGESSDAAPSADAAQ | |||||||||||||||||||
| 10 | 4ww3A | 0.14 | 0.18 | 0.94 | 3.11 | Download | ETWWYNPSIVVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKT-KSLQTPAN----MFIINLAFSDFTFSLVNGFPLMTISC---FLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPL-EWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETEDDKDAET-------EIPAGE----- | |||||||||||||||||||
| ||||||||||||||||||||||||||
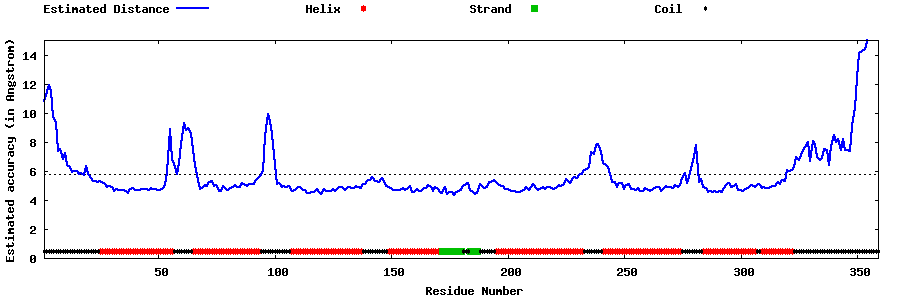
| Generated 3D models | Estimated local accuracy of models | ||
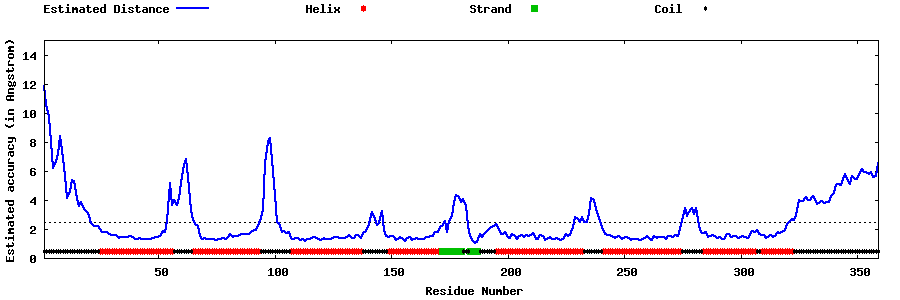 | |||
|
|
|||
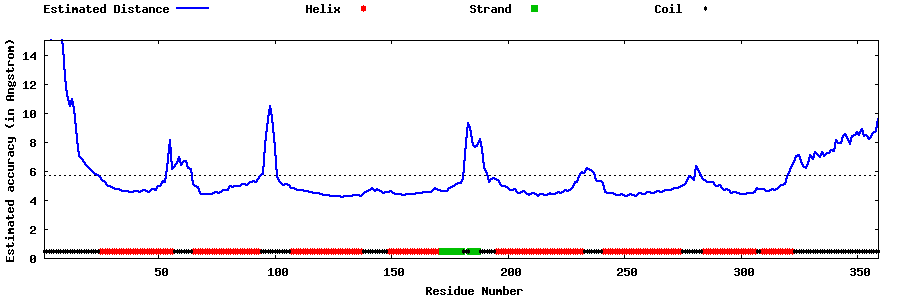 | |||
|
| |||
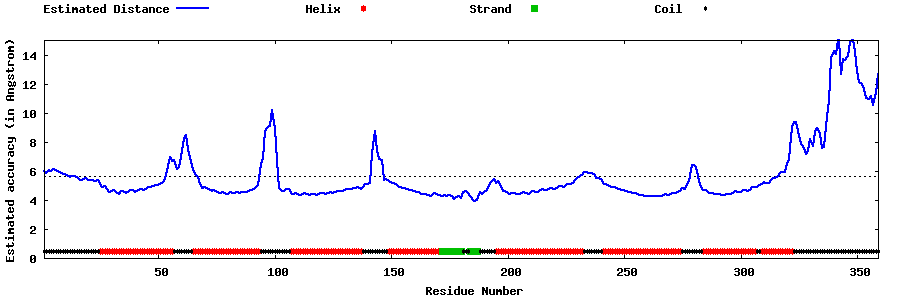 | |||
|
| |||
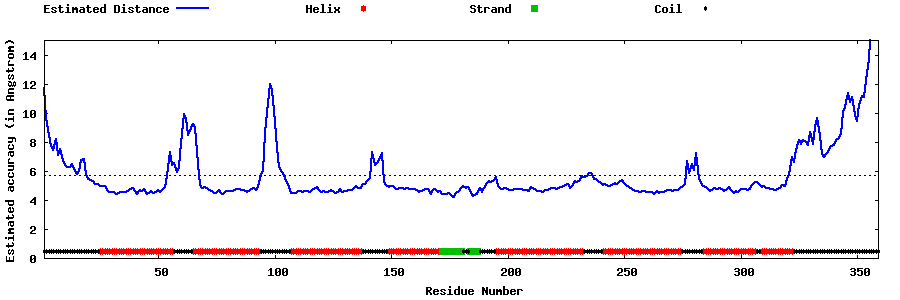 | |||
|
| |||
 | |||
|
| |||