

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MGNASNDSQSEDCETRQWLPPGESPAISSVMFSAGVLGNLIALALLARRWRGDVGCSAGRRSSLSLFHVLVTELVFTDLLGTCLISPVVLASYARNQTLVALAPESRACTYFAFAMTFFSLATMLMLFAMALERYLSIGHPYFYQRRVSRSGGLAVLPVIYAVSLLFCSLPLLDYGQYVQYCPGTWCFIRHGRTAYLQLYATLLLLLIVSVLACNFSVILNLIRMHRRSRRSRCGPSLGSGRGGPGARRRGERVSMAEETDHLILLAIMTITFAVCSLPFTIFAYMNETSSRKEKWDLQALRFLSINSIIDPWVFAILRPPVLRLMRSVLCCRISLRTQDATQTSCSTQSDASKQADL | |
| CCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCCCCCCSSSCCCCCHHHHHHHSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9999899888888776644104999999999999999999988877650443211100132157779999999999999999998899999998578514778988965629999999999999999999997386545546177758588999999999999999999998740422411897456165775056664303431022578999999999999999999875225544555677543332033688999999999999999999999999999999974778861999999999999999899999439999999999948889998898877888788987557899 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MGNASNDSQSEDCETRQWLPPGESPAISSVMFSAGVLGNLIALALLARRWRGDVGCSAGRRSSLSLFHVLVTELVFTDLLGTCLISPVVLASYARNQTLVALAPESRACTYFAFAMTFFSLATMLMLFAMALERYLSIGHPYFYQRRVSRSGGLAVLPVIYAVSLLFCSLPLLDYGQYVQYCPGTWCFIRHGRTAYLQLYATLLLLLIVSVLACNFSVILNLIRMHRRSRRSRCGPSLGSGRGGPGARRRGERVSMAEETDHLILLAIMTITFAVCSLPFTIFAYMNETSSRKEKWDLQALRFLSINSIIDPWVFAILRPPVLRLMRSVLCCRISLRTQDATQTSCSTQSDASKQADL | |
| 7524334344442534432322010101121133133112100000001233333334344323000000000001001210220100000010232000012322200100000001002000100000000000000000204430232000000000002001002101100012243342000011132200010102323331222012001000000122243344444444444444444443443333330120000000001000000111000000021232432000000330320002100140001340040024101022324446355443344443345456 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCCCCCCSSSCCCCCHHHHHHHSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCC MGNASNDSQSEDCETRQWLPPGESPAISSVMFSAGVLGNLIALALLARRWRGDVGCSAGRRSSLSLFHVLVTELVFTDLLGTCLISPVVLASYARNQTLVALAPESRACTYFAFAMTFFSLATMLMLFAMALERYLSIGHPYFYQRRVSRSGGLAVLPVIYAVSLLFCSLPLLDYGQYVQYCPGTWCFIRHGRTAYLQLYATLLLLLIVSVLACNFSVILNLIRMHRRSRRSRCGPSLGSGRGGPGARRRGERVSMAEETDHLILLAIMTITFAVCSLPFTIFAYMNETSSRKEKWDLQALRFLSINSIIDPWVFAILRPPVLRLMRSVLCCRISLRTQDATQTSCSTQSDASKQADL | |||||||||||||||||||||||||
| 1 | 5tjvA | 0.16 | 0.19 | 0.80 | 2.71 | Download | ----ENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHS-----------RSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSR-----NVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNC---EKLQSVCSDIFPHDETYLMFWIGVTSVLLLFIVYAYMYILWKA----------------------GKRAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF---------------------------- | |||||||||||||||||||
| 2 | 4amjA | 0.19 | 0.20 | 0.78 | 3.82 | Download | --------------------EAGMSLLMALVVLLIVAGNVLVIAAIGST-----------QRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWG----SFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKE-------------QIRKIDRASKRKTSRVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRVPDWLFVAFNWLGYANSAMNPIIYCR-SPDFRKAFKRLL---------------------------- | |||||||||||||||||||
| 3 | 4iaqA | 0.16 | 0.19 | 0.78 | 3.09 | Download | ---------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRT-----------RKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTL----GQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPP-------FFWRQASECVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSR---------------------IIQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVM---PIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------------------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.14 | 0.17 | 0.85 | 1.54 | Download | --------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKL-----------QYATNYFLMSLAVADLLVGLFVMPIALLTIMFEAMW---PLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIGIET---NPNNITCVLTKERGDFMLGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETADNAQKGDGQNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR----------------------- | |||||||||||||||||||
| 5 | 4ib4 | 0.13 | 0.17 | 0.85 | 1.18 | Download | --------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLE-----------KKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFAMWPLP----LVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPPIKGIETN---PNNITCVLTKERGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDDQKGQKNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR----------------------- | |||||||||||||||||||
| 6 | 4iaqA | 0.16 | 0.19 | 0.77 | 2.85 | Download | ------------QDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRT-----------RKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLG----QVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFWRQASE-------CVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRI---------------------IQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPI---HLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------------------------- | |||||||||||||||||||
| 7 | 3uon | 0.12 | 0.20 | 0.82 | 1.72 | Download | ----------------------FIVLVAGSLSLVTIIGNILVMVSIKVN-----------RHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVI----GYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGTVEDGECYIQFFSNAVTFGTAIAAFYLPVIMTVLYWHISRASKSRINIFEMEGGHSLNAAIGNAFRTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM--------------------------- | |||||||||||||||||||
| 8 | 1u19A | 0.15 | 0.18 | 0.90 | 4.64 | Download | FSNKTGVVRSPFEAPQYYLAESMLAAYMFLLIMLGFPINFLTLYVTVQH-----------KKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVF----GPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPM-SNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYNNESFVIYMFVVHFIIPLIVIFFCYG-------------------QLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEASTTVSKTETSQVAPA- | |||||||||||||||||||
| 9 | 4iaqA | 0.16 | 0.19 | 0.78 | 2.73 | Download | ---------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRT-----------RKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLG----QVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFWRQASE-------CVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRI---------------------IQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMP---IHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------------------------- | |||||||||||||||||||
| 10 | 3zpqA | 0.19 | 0.19 | 0.78 | 3.19 | Download | -----------GAELLSQQWEAGMSLLMALVVLLIVAGNVLVIAAIG-----------STQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWG----SFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQ-------------------------SRVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRVPDWLFVAFNWLGYANSAMNPIIYCR-SPDFRKAFKRLLA--------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
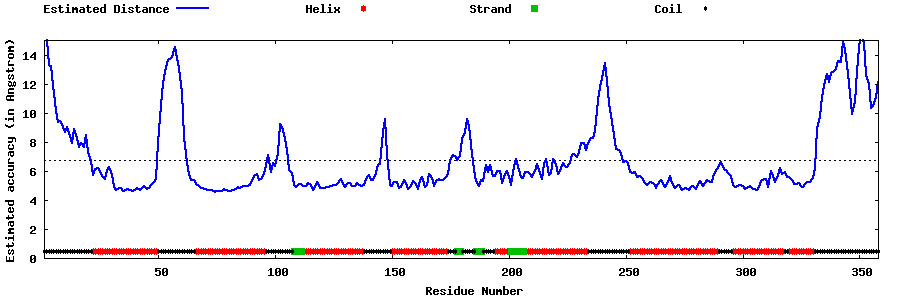
| Generated 3D models | Estimated local accuracy of models | ||
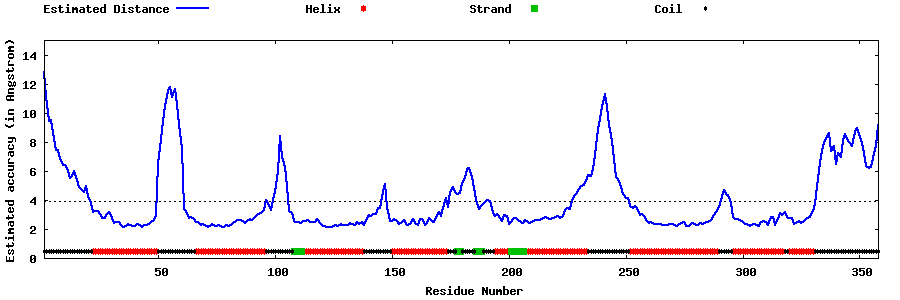 | |||
|
|
|||
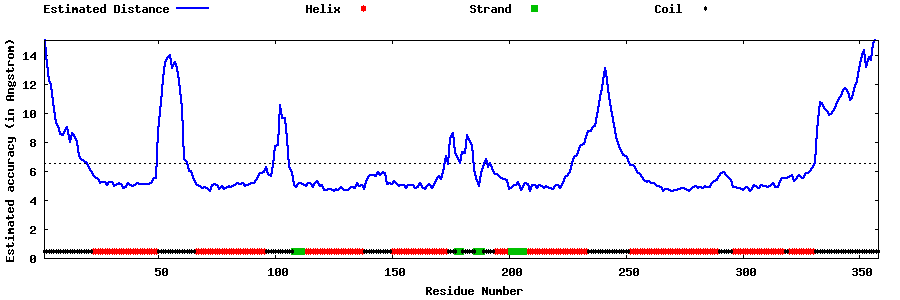 | |||
|
| |||
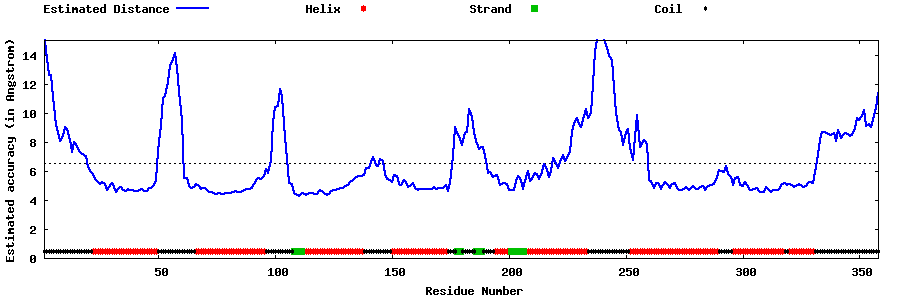 | |||
|
| |||
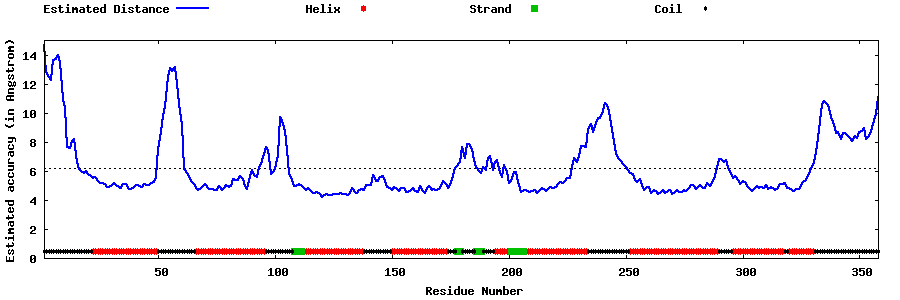 | |||
|
| |||
 | |||
|
| |||