

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | |
| | | | | | | | | | | | | | | | |
| MITFLYIFFSILIMVLFVLGNFANGFIALVNFIDWVKRKKISSADQILTALAVSRIGLLWALLLNWYLTVLNPAFYSVELRITSYNAWVVTNHFSMWLAANLSIFYLLKIANFSNLLFLHLKRRVRSVILVILLGTLIFLVCHLLVANMDESMWAEEYEGNMTGKMKLRNTVHLSYLTVTTLWSFIPFTLSLISFLMLICSLCKHLKKMQLHGEGSQDLSTKVHIKALQTLISFLLLCAIFFLFLIVSVWSPRRLRNDPVVMVSKAVGNIYLAFDSFILIWRTKKLKHTFLLILCQIRC | |
| CCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCSSSSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHCCC | |
| 96489999999999999999999899999999999827988777699999999999999999972655001520113236999988999983899999999999982464378977999999865270999999999999999999970022215778883205665002488999999999999999999999999999999999996087999999981999999999999999999999999999998730333899999999999837887699960870999999999976679 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | |
| | | | | | | | | | | | | | | | |
| MITFLYIFFSILIMVLFVLGNFANGFIALVNFIDWVKRKKISSADQILTALAVSRIGLLWALLLNWYLTVLNPAFYSVELRITSYNAWVVTNHFSMWLAANLSIFYLLKIANFSNLLFLHLKRRVRSVILVILLGTLIFLVCHLLVANMDESMWAEEYEGNMTGKMKLRNTVHLSYLTVTTLWSFIPFTLSLISFLMLICSLCKHLKKMQLHGEGSQDLSTKVHIKALQTLISFLLLCAIFFLFLIVSVWSPRRLRNDPVVMVSKAVGNIYLAFDSFILIWRTKKLKHTFLLILCQIRC | |
| 73333332113213312331331022002000110045340320110000000010100121333100000102123330000010002221230010000001000000022311000001330421001103312233332213120333132643421211313133322011112333333323311331332021002202320243344331130400220010022122323323311331111233343310111111200210331020302114301300130023036 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCSSSSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHCCC MITFLYIFFSILIMVLFVLGNFANGFIALVNFIDWVKRKKISSADQILTALAVSRIGLLWALLLNWYLTVLNPAFYSVELRITSYNAWVVTNHFSMWLAANLSIFYLLKIANFSNLLFLHLKRRVRSVILVILLGTLIFLVCHLLVANMDESMWAEEYEGNMTGKMKLRNTVHLSYLTVTTLWSFIPFTLSLISFLMLICSLCKHLKKMQLHGEGSQDLSTKVHIKALQTLISFLLLCAIFFLFLIVSVWSPRRLRNDPVVMVSKAVGNIYLAFDSFILIWRTKKLKHTFLLILCQIRC | |||||||||||||||||||||||||
| 1 | 4djhA | 0.10 | 0.21 | 0.92 | 1.41 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR----TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKA----LDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSDR-----------NLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--- | |||||||||||||||||||
| 2 | 5tjvA | 0.10 | 0.20 | 0.92 | 2.65 | Download | LNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHS--RSLRCRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPK----AVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIF-----PHIDETYLMFWIGVTSVLLLFIVYAYMYILWKAGKRAMSFSD--------QARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----- | |||||||||||||||||||
| 3 | 4djhA | 0.10 | 0.21 | 0.92 | 2.45 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRY--TKMK-TATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKAL----DFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLL-------------RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--- | |||||||||||||||||||
| 4 | 4djh | 0.08 | 0.15 | 0.95 | 1.56 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRY---TKMKTATNIYIFNLALADALVTTT-MPFQSTVYLMNSWPFGDVLCIVLSIDYYNMFTSIFTLTMMSVDRYIAVC---HPVKAL-DFRTPLKAKIINICIWLLSSVGISAIVLGGKVDDVECSLQFPDDDYWWDLFMKI--CVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFTDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--- | |||||||||||||||||||
| 5 | 3odu | 0.16 | 0.21 | 0.97 | 1.29 | Download | NANFNKIFLPTIYSIIFLTGIVGNGLVILVMGY---QKKLRSMTDKYRLHLSVADLLFVIT-LPFWAVDAVANWYFGNFLCKAVHVIYTVNLYSSVWILAFISLDRYLAIVHATNSQ----RPRKLLAEKVVYVGVWIPALLLTIPDFIFANVSEADDRYICDRFYPNDLWVVVFQFQHIMVGLILPGIVILSCYCIIISKLSHSGSNIFEMLRDAYGSKGHQKRKALKTTVILILAFFACWLPYYIGISISFILLVHKWISITEALAFFHCCLNPILYAFLGAKFKTSAQHALTSGRP | |||||||||||||||||||
| 6 | 4djhA | 0.10 | 0.21 | 0.93 | 1.56 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRY---TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKA----LDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSD-----------RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--- | |||||||||||||||||||
| 7 | 4djh | 0.10 | 0.15 | 0.96 | 1.70 | Download | --PAIPVIITAVYSVVFVVGLVGNSLVMFVIIR---YTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVD--VIECSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS---AALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--- | |||||||||||||||||||
| 8 | 4buoA | 0.15 | 0.22 | 0.94 | 2.19 | Download | TDIYSKVLVTAIYLALFVVGTVGNSVTLFTLAR---KKSLQSTVDYYLGSLALSDLLILLLVELYNFIWVHHPWAFGDAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIP--MLFTMGLQNLSGDGTHPGGLVCTPIATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQ---------PGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL----- | |||||||||||||||||||
| 9 | 4djhA | 0.10 | 0.21 | 0.92 | 1.46 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR----TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVC---HPVKA-LDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSD-----------RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--- | |||||||||||||||||||
| 10 | 3zevA | 0.14 | 0.22 | 0.95 | 1.61 | Download | TDIYSKVLVTAIYLALFVVGTVGNSVTLFTLA----RKKSLSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHFGDAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGLVCTPIVTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVH---------QPGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLST---LAC | |||||||||||||||||||
| ||||||||||||||||||||||||||
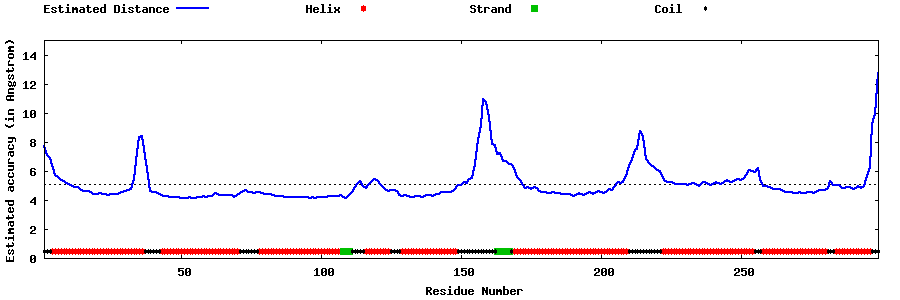
| Generated 3D models | Estimated local accuracy of models | ||
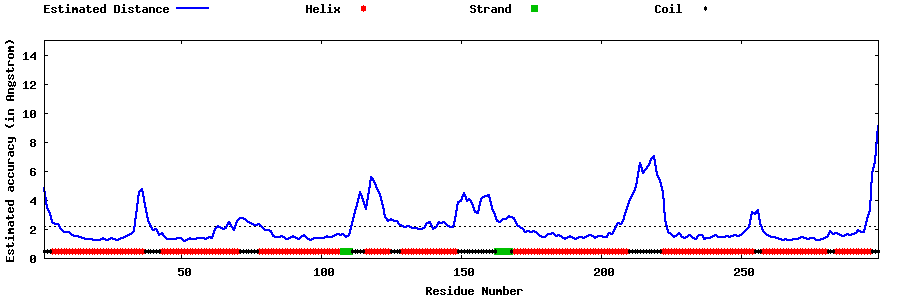 | |||
|
|
|||
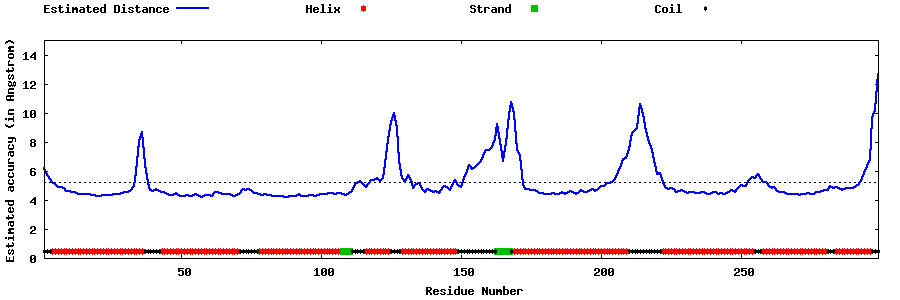 | |||
|
| |||
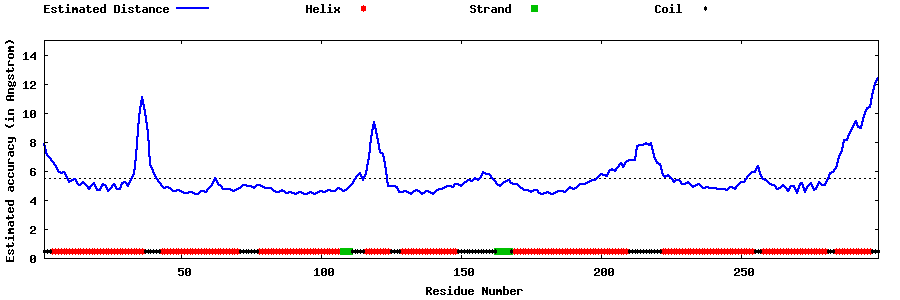 | |||
|
| |||
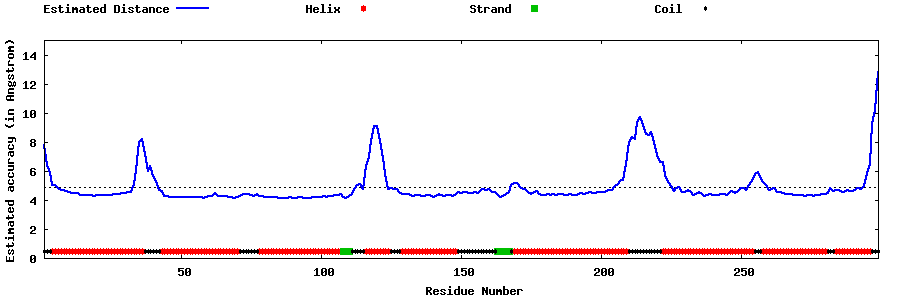 | |||
|
| |||
 | |||
|
| |||