

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MNTTSSAAPPSLGVEFISLLAIILLSVALAVGLPGNSFVVWSILKRMQKRSVTALMVLNLALADLAVLLTAPFFLHFLAQGTWSFGLAGCRLCHYVCGVSMYASVLLITAMSLDRSLAVARPFVSQKLRTKAMARRVLAGIWVLSFLLATPVLAYRTVVPWKTNMSLCFPRYPSEGHRAFHLIFEAVTGFLLPFLAVVASYSDIGRRLQARRFRRSRRTGRLVVLIILTFAAFWLPYHVVNLAEAGRALAGQAAGLGLVGKRLSLARNVLIALAFLSSSVNPVLYACAGGGLLRSAGVGFVAKLLEGTGSEASSTRRGGSLGQTARSGPAALEPGPSESLTASSPLKLNELN | |
| CCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9999998988732419999999999999999988799823522048877869999999999999999995899999995197889656898999999999999999999999971788742053100216788999999999999999969999853477899936861889945899999999999999999999999999999999754452465489999999999999989999999999999843566523678999999999999999999848589899619889999999987985356566677877888877777888877889999987849998646699 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MNTTSSAAPPSLGVEFISLLAIILLSVALAVGLPGNSFVVWSILKRMQKRSVTALMVLNLALADLAVLLTAPFFLHFLAQGTWSFGLAGCRLCHYVCGVSMYASVLLITAMSLDRSLAVARPFVSQKLRTKAMARRVLAGIWVLSFLLATPVLAYRTVVPWKTNMSLCFPRYPSEGHRAFHLIFEAVTGFLLPFLAVVASYSDIGRRLQARRFRRSRRTGRLVVLIILTFAAFWLPYHVVNLAEAGRALAGQAAGLGLVGKRLSLARNVLIALAFLSSSVNPVLYACAGGGLLRSAGVGFVAKLLEGTGSEASSTRRGGSLGQTARSGPAALEPGPSESLTASSPLKLNELN | |
| 7143443135633230100000210120022033232201100011333210000000000100000000000000100363310020011001000300110000000000100000000002024213211010000001100000010000002024376321101030237202200000111201221120002000000210243445533000000000000010031211000001002303333342332331020010000000030002102020000330142004001220244345644434444344444544444445443641446341646638 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MNTTSSAAPPSLGVEFISLLAIILLSVALAVGLPGNSFVVWSILKRMQKRSVTALMVLNLALADLAVLLTAPFFLHFLAQGTWSFGLAGCRLCHYVCGVSMYASVLLITAMSLDRSLAVARPFVSQKLRTKAMARRVLAGIWVLSFLLATPVLAYRTVVPWKTNMSLCFPRYPSEGHRAFHLIFEAVTGFLLPFLAVVASYSDIGRRLQARRFRRSRRTGRLVVLIILTFAAFWLPYHVVNLAEAGRALAGQAAGLGLVGKRLSLARNVLIALAFLSSSVNPVLYACAGGGLLRSAGVGFVAKLLEGTGSEASSTRRGGSLGQTARSGPAALEPGPSESLTASSPLKLNELN | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.25 | 0.21 | 0.83 | 3.32 | Download | ------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA-QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGL-HYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQ--EFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ--------------------------------------------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.26 | 0.21 | 0.82 | 4.06 | Download | ------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA-AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQK-EGLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNN--CSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFF---------------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.27 | 0.24 | 0.83 | 3.87 | Download | -----SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV-------DIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------------------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.18 | 0.23 | 0.95 | 1.51 | Download | YNPSIIVHPFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEANAEMRLAKISIVIVSQFLLSWSPYAVVALLA----QFGPLE------WVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETED-DK--D----AETEIPAGESSDAAPSADAAQMKE-- | |||||||||||||||||||
| 5 | 4djh | 0.29 | 0.24 | 0.78 | 1.17 | Download | -------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS----A---------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP-------------------------------------------------- | |||||||||||||||||||
| 6 | 4mbsA | 0.25 | 0.21 | 0.82 | 3.33 | Download | -------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA-QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQK-EGLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEF--FGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ--------------------------------------------------- | |||||||||||||||||||
| 7 | 4djh | 0.29 | 0.24 | 0.77 | 1.72 | Download | ----------------IPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDYDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS----A---------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF--------------------------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.26 | 0.22 | 0.82 | 4.75 | Download | ------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA-QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSFPYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREKKRHRD--VRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNN--CSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ--------------------------------------------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.25 | 0.21 | 0.83 | 3.44 | Download | ------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA-AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQK-EGLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEKHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQ--EFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ--------------------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.27 | 0.23 | 0.81 | 4.56 | Download | --GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKY-RQGSIDCTLTFPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPET--------TFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF-------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
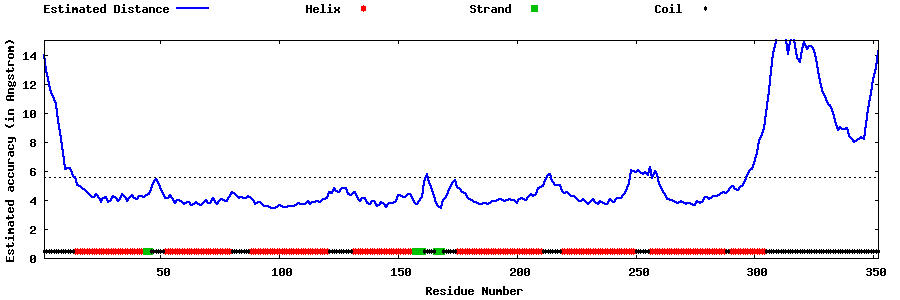
| Generated 3D models | Estimated local accuracy of models | ||
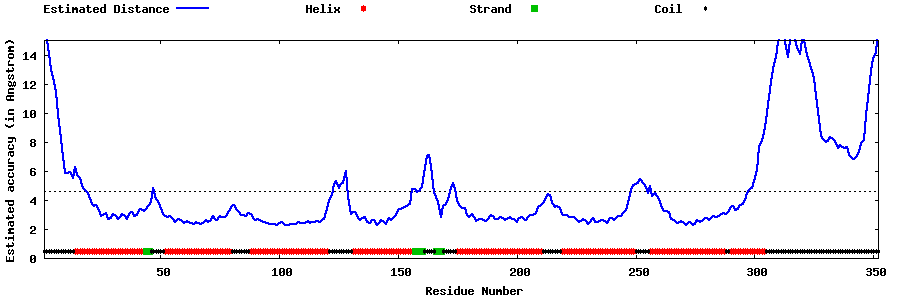 | |||
|
|
|||
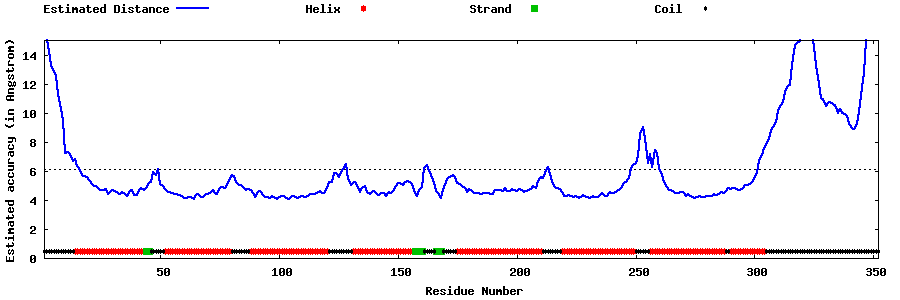 | |||
|
| |||
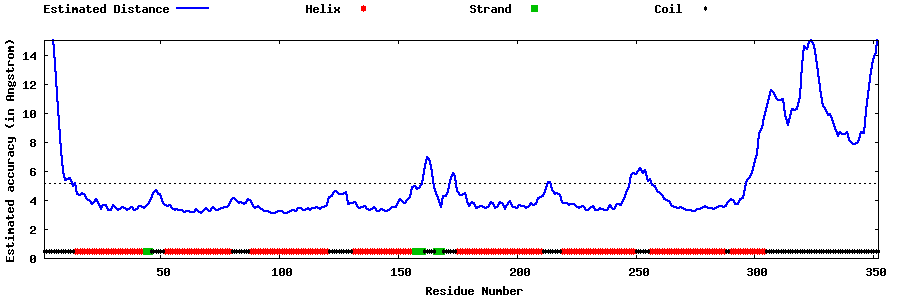 | |||
|
| |||
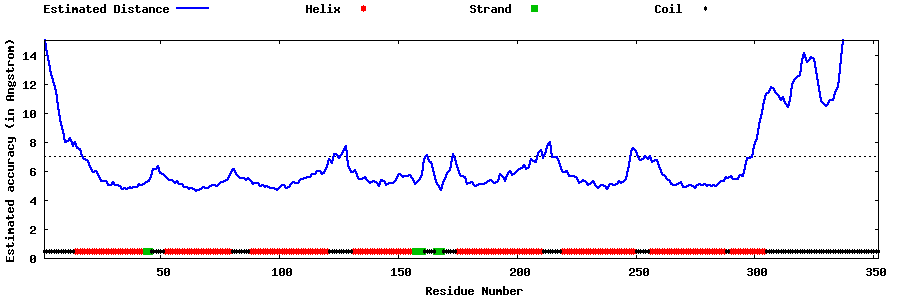 | |||
|
| |||
 | |||
|
| |||