

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MPTVNHSGTSHTVFHLLGIPGLQDQHMWISIPFFISYVTALLGNSLLIFIILTKRSLHEPMYLFLCMLAGADIVLSTCTIPQALAIFWFRAGDISLDRCITQLFFIHSTFISESGILLVMAFDHYIAICYPLRYTTILTNALIKKICVTVSLRSYGTIFPIIFLLKRLTFCQNNIIPHTFCEHIGLAKYACNDIRINIWYGFSILMSTVVLDVVLIFISYMLILHAVFHMPSPDACHKALNTFGSHVCIIILFYGSGIFTILTQRFGRHIPPCIHIPLANVCILAPPMLNPIIYGIKTKQIQEQVVQFLFIKQK | |
| CCCCCCCCCCCHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHCCCCC | |
| 99988998804013057998986789999999999999999999999999856887543089999999999999999669999999966998578888999999999999999999999982427740666556312888999999999999999999889999952899999935872211376788835784475799999999999999999999999999998178897688898550599999999999999998636414898884899999999983886646812215479999999998333779 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MPTVNHSGTSHTVFHLLGIPGLQDQHMWISIPFFISYVTALLGNSLLIFIILTKRSLHEPMYLFLCMLAGADIVLSTCTIPQALAIFWFRAGDISLDRCITQLFFIHSTFISESGILLVMAFDHYIAICYPLRYTTILTNALIKKICVTVSLRSYGTIFPIIFLLKRLTFCQNNIIPHTFCEHIGLAKYACNDIRINIWYGFSILMSTVVLDVVLIFISYMLILHAVFHMPSPDACHKALNTFGSHVCIIILFYGSGIFTILTQRFGRHIPPCIHIPLANVCILAPPMLNPIIYGIKTKQIQEQVVQFLFIKQK | |
| 54442424231310200000121320200011022012203321210000020151001000200010020000002000020000000204403040000001200310230100011002000000021010000003410010000002212011202102021032045200000001021001000230300011002003312331231032011200200020014601320120010010001003231331010010033002100010022100301330030000203400410030024468 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHCCCCC MPTVNHSGTSHTVFHLLGIPGLQDQHMWISIPFFISYVTALLGNSLLIFIILTKRSLHEPMYLFLCMLAGADIVLSTCTIPQALAIFWFRAGDISLDRCITQLFFIHSTFISESGILLVMAFDHYIAICYPLRYTTILTNALIKKICVTVSLRSYGTIFPIIFLLKRLTFCQNNIIPHTFCEHIGLAKYACNDIRINIWYGFSILMSTVVLDVVLIFISYMLILHAVFHMPSPDACHKALNTFGSHVCIIILFYGSGIFTILTQRFGRHIPPCIHIPLANVCILAPPMLNPIIYGIKTKQIQEQVVQFLFIKQK | |||||||||||||||||||||||||
| 1 | 3emlA | 0.20 | 0.24 | 0.88 | 2.46 | Download | ---------------------IM-GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACL-----------FEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC---PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHRQ | |||||||||||||||||||
| 2 | 5tgzA | 0.20 | 0.23 | 0.86 | 2.07 | Download | -----------------------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-------GWNCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDRMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLLLNSTVNPIIYALRSKDLRHAFRSMF----- | |||||||||||||||||||
| 3 | 5tgzA | 0.20 | 0.23 | 0.88 | 1.79 | Download | FMDIECFMVLNP-----------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFH-VFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLNCEKLQSVCSDIFPHID--------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVF-GKMNIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----- | |||||||||||||||||||
| 4 | 3uon | 0.17 | 0.15 | 0.88 | 1.55 | Download | ----------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIV--GVRTVEDGECYIQFF------S---NAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINISREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPIPNTV-WTIGYWLCYINSTINPACYALCNATFKKTFKHLLM---- | |||||||||||||||||||
| 5 | 3uonA | 0.16 | 0.17 | 0.87 | 1.13 | Download | ----------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQF--IVGVRTVEDGECYIQFF---------SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKS----RREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM---- | |||||||||||||||||||
| 6 | 3emlA | 0.20 | 0.24 | 0.88 | 2.64 | Download | -----------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVV-----------PMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC---PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 7 | 4iaq | 0.18 | 0.21 | 0.83 | 1.71 | Download | -----------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQASE----------CVVN----------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLG---IILGAFIVCWLIISLVMPIH----LAIF-DFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK-- | |||||||||||||||||||
| 8 | 4buoA | 0.14 | 0.18 | 0.94 | 2.70 | Download | ----NSD---------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGLVCTPIVDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDEQWTYHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL----- | |||||||||||||||||||
| 9 | 5tgzA | 0.20 | 0.23 | 0.90 | 3.13 | Download | GRGENFMDIEC--FMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG--------NCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKA-APDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----- | |||||||||||||||||||
| 10 | 4gpoA | 0.21 | 0.24 | 0.89 | 4.74 | Download | ---------------------LQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPG--------CCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYC-RSPDFRKAFKRL------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
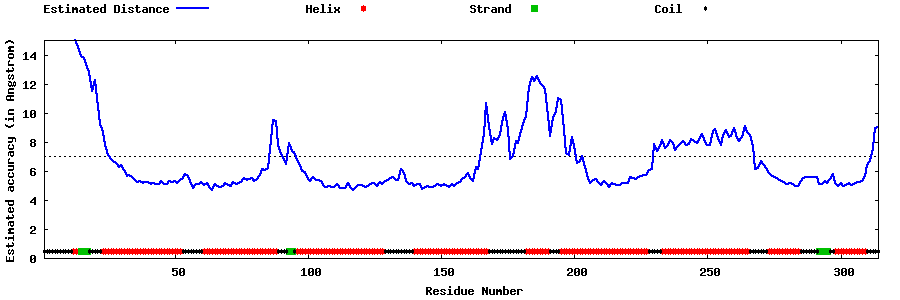
| Generated 3D models | Estimated local accuracy of models | ||
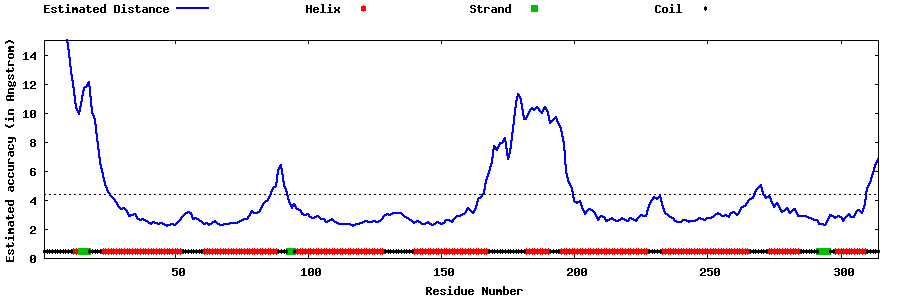 | |||
|
|
|||
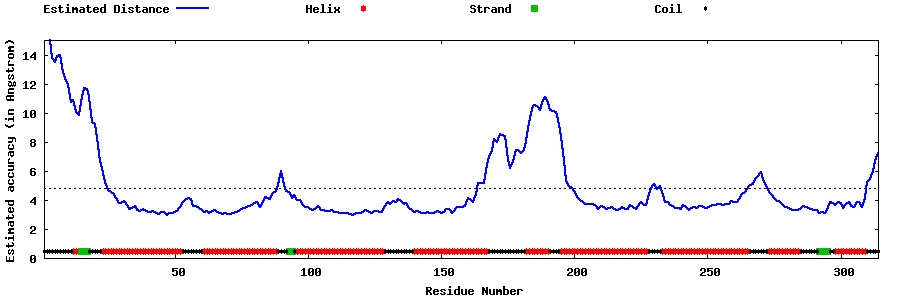 | |||
|
| |||
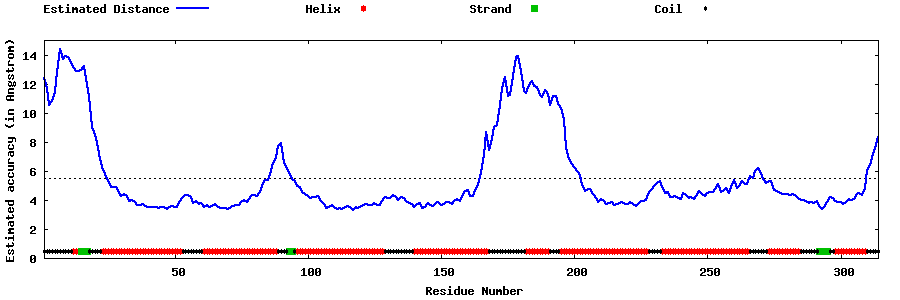 | |||
|
| |||
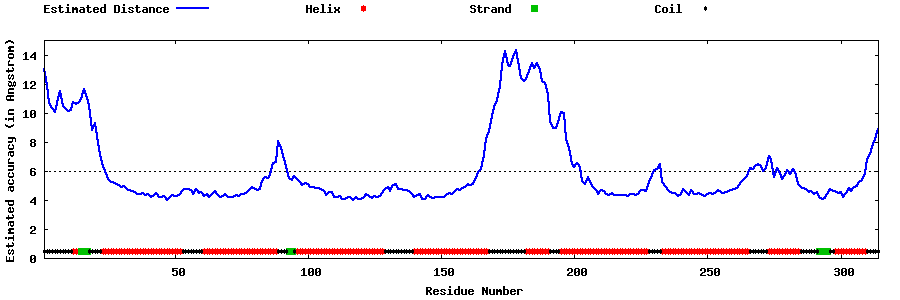 | |||
|
| |||
 | |||
|
| |||