

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MQLNNNVTEFILLGLTQDPFWKKIVFVIFLRLYLGTLLGNLLIIISVKTSQALKNPMFFFLFYLSLSDTCLSTSITPRMIVDALLKKTTISFSECMIQVFSSHVFGCLEIFILILTAVDRYVDICKPLHYMTIISQWVCGVLMAVAWVGSCVHSLVQIFLALSLPFCGPNVINHCFCDLQPLLKQACSETYVVNLLLVSNSGAICAVSYVMLIFSYVIFLHSLRNHSAEVIKKALSTCVSHIIVVILFFGPCIFMYTCLATVFPMDKMIAVFYTVGTSFLNPVIYTLKNTEVKSAMRKLWSKKLITDDKR | |
| CCCCCCSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHCCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCSSCCCHHHHHHHHCCCCHHCSSHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHCSSSSSCCCCCCCCCCCSSHHHHHCHHHHCCCHHHCCCCHHHHHHHHHHHHHHCCCCCCC | |
| 9999732134661599980247999999999999999978875551221799999789998869987642005676898998751896786899999999999999989999999986528863611016102677599999999999999999999999624788898847776428189887830241522011100123999999999999999999985245731211898888879992646620643688588999965001313454402300036432461999999999997303545689 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MQLNNNVTEFILLGLTQDPFWKKIVFVIFLRLYLGTLLGNLLIIISVKTSQALKNPMFFFLFYLSLSDTCLSTSITPRMIVDALLKKTTISFSECMIQVFSSHVFGCLEIFILILTAVDRYVDICKPLHYMTIISQWVCGVLMAVAWVGSCVHSLVQIFLALSLPFCGPNVINHCFCDLQPLLKQACSETYVVNLLLVSNSGAICAVSYVMLIFSYVIFLHSLRNHSAEVIKKALSTCVSHIIVVILFFGPCIFMYTCLATVFPMDKMIAVFYTVGTSFLNPVIYTLKNTEVKSAMRKLWSKKLITDDKR | |
| 8756220000000000424500100013133323323332330000020043030000100230012001100000020000002532100050001000110331131020002003000000022110100013310000012013102301300131023030001022000100222003000020230003002301320231033133112100100132146122201100300210020132101000000333133000100310231033003000120440230033013430325668 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHCCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCSSCCCHHHHHHHHCCCCHHCSSHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHCSSSSSCCCCCCCCCCCSSHHHHHCHHHHCCCHHHCCCCHHHHHHHHHHHHHHCCCCCCC MQLNNNVTEFILLGLTQDPFWKKIVFVIFLRLYLGTLLGNLLIIISVKTSQALKNPMFFFLFYLSLSDTCLSTSITPRMIVDALLKKTTISFSECMIQVFSSHVFGCLEIFILILTAVDRYVDICKPLHYMTIISQWVCGVLMAVAWVGSCVHSLVQIFLALSLPFCGPNVINHCFCDLQPLLKQACSETYVVNLLLVSNSGAICAVSYVMLIFSYVIFLHSLRNHSAEVIKKALSTCVSHIIVVILFFGPCIFMYTCLATVFPMDKMIAVFYTVGTSFLNPVIYTLKNTEVKSAMRKLWSKKLITDDKR | |||||||||||||||||||||||||
| 1 | 3emlA | 0.19 | 0.24 | 0.88 | 3.42 | Download | ------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVPMNYMVYFN-----------FFACVLVPLLLMLGVYLRIFLAARQLVHAAKSLAIIVGLFALCWLPLHII-NCFTFFCPDCSHAWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ--- | |||||||||||||||||||
| 2 | 5tgzA | 0.20 | 0.23 | 0.89 | 2.14 | Download | -ENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL--------GWNCEK---------LQSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARELAKTLVLILVVLIICWGPLLAIMVKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSM----------- | |||||||||||||||||||
| 3 | 5tgzA | 0.19 | 0.23 | 0.90 | 2.09 | Download | -GGRGENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFH-VFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLCEKLQSVC-------------------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHQARMDIELAKTLVLILVVLIICWGPLLAIMVGKMNLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF---------- | |||||||||||||||||||
| 4 | 4djh | 0.16 | 0.22 | 0.90 | 1.53 | Download | -----------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP-------- | |||||||||||||||||||
| 5 | 4yay | 0.16 | 0.21 | 0.90 | 1.21 | Download | KTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK-----KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL---------- | |||||||||||||||||||
| 6 | 3emlA | 0.20 | 0.24 | 0.89 | 3.56 | Download | -------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFCPDCSHAPLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ--- | |||||||||||||||||||
| 7 | 4iaq | 0.22 | 0.23 | 0.85 | 1.73 | Download | ------------------LPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASECV----------VNT-------D---HILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADRERKATKTLGIILGAFIVCWLPFFIISLVMPI-HLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------- | |||||||||||||||||||
| 8 | 4buoA | 0.16 | 0.20 | 0.88 | 2.67 | Download | ---NSD------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGLVCTPIV------DTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQAVVIAFVVCWLPYHVRRLMFCYI-----------SDEQMLTNALVYVSAAINPILYNLVSANFRQVFLSTL---------- | |||||||||||||||||||
| 9 | 5tgzA | 0.20 | 0.23 | 0.89 | 4.78 | Download | GENFMDIECFMVL----NPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVF-HRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG---------NCEK---------LQSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQAIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMN-FAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF---------- | |||||||||||||||||||
| 10 | 2ydoA | 0.18 | 0.22 | 0.93 | 5.50 | Download | ---------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRLKQMESTLQKEVHAAKSLAIIVGLFALCCFTFFCPDCSHAPLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQEP | |||||||||||||||||||
| ||||||||||||||||||||||||||
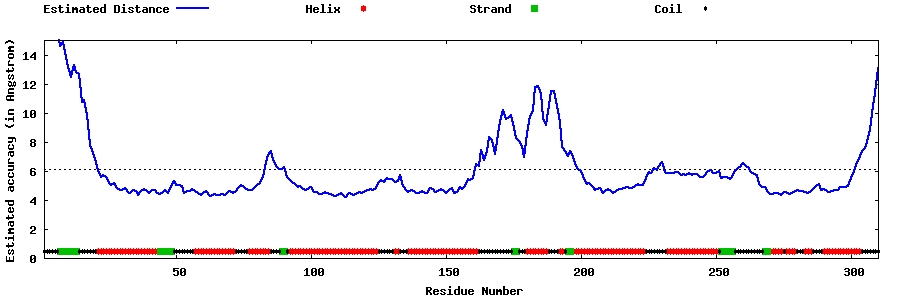
| Generated 3D models | Estimated local accuracy of models | ||
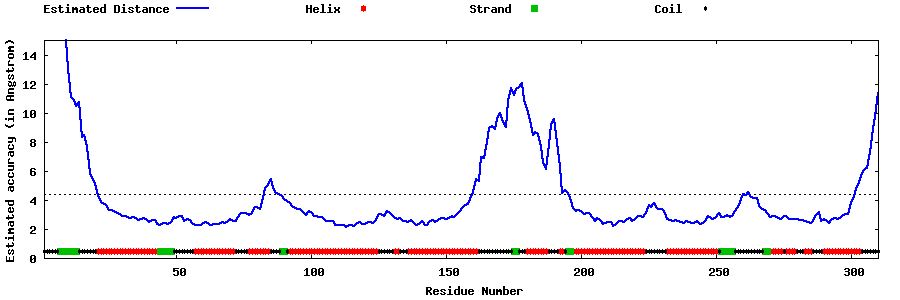 | |||
|
|
|||
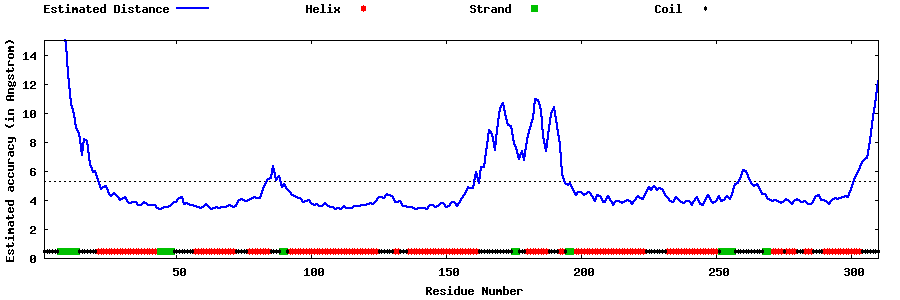 | |||
|
| |||
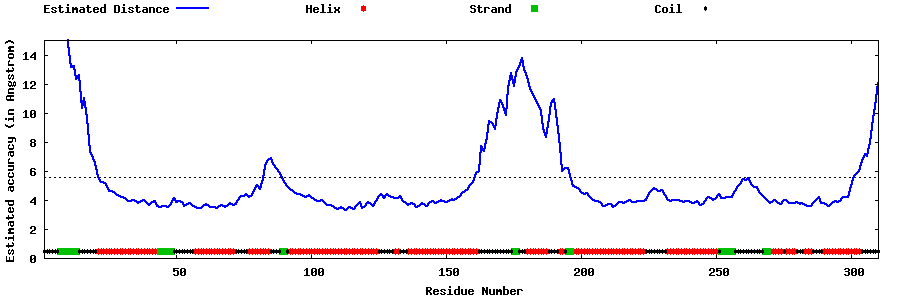 | |||
|
| |||
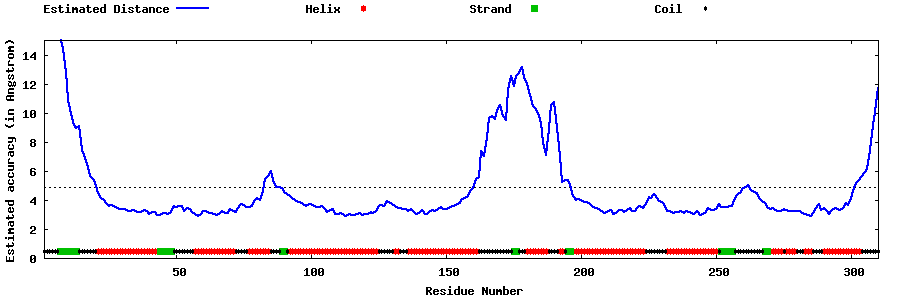 | |||
|
| |||
 | |||
|
| |||