

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MDKENSSMVTEFIFMGITQDPQMEIIFFVVFLIVYLVNVVGNIGMIILITTDTQLHTPMYFFLCNLSFVDLGYSSAIAPRMLADFLTNHKVISFSSCATQFAFFVGFVDAECYVLAAMAYGRFVAICRPLHYSTFMSKQVCLALMLGSYLAGLVSLVAHTTLTFSLSYCGSNIINHFFCEIPPLLALSCSDTYISEILLFSLCGFIEFSTILIIFISYTFILVAIIRMRSAEGRLKAFSTCGSHLTGITLFYGTVMFMYLRPTSSYSLDQDKWASVFYTVIIPMLNPLIYSLRNKDVKAAFKKLIGKKSQ | |
| CCCCCCCCCSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHCCHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCSSSSSSCCHHHCCCCHHHCCCCHHHHHHHHHHHHHHCC | |
| 9998884230236864789925289999999999999999998999999618887774888876799884472114619999987148978858999999999999999999999999865145206110188026788999999999999999999999998453089989048641681888877156862767888989999999999999999999999980266767675420164888997999974330348378999888789789988221031233574404649999999999974269 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MDKENSSMVTEFIFMGITQDPQMEIIFFVVFLIVYLVNVVGNIGMIILITTDTQLHTPMYFFLCNLSFVDLGYSSAIAPRMLADFLTNHKVISFSSCATQFAFFVGFVDAECYVLAAMAYGRFVAICRPLHYSTFMSKQVCLALMLGSYLAGLVSLVAHTTLTFSLSYCGSNIINHFFCEIPPLLALSCSDTYISEILLFSLCGFIEFSTILIIFISYTFILVAIIRMRSAEGRLKAFSTCGSHLTGITLFYGTVMFMYLRPTSSYSLDQDKWASVFYTVIIPMLNPLIYSLRNKDVKAAFKKLIGKKSQ | |
| 8666230200100000004325010100130333133133333101200200130000001002000000010000000300010015422010300010010000001000000000000000000201101030033000000120011013001010000020201432201000001200020001323010100010013223303300330331001000103026323101000110200010232012000000324312344200002103200230030000213302400330154358 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHCCHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCSSSSSSCCHHHCCCCHHHCCCCHHHHHHHHHHHHHHCC MDKENSSMVTEFIFMGITQDPQMEIIFFVVFLIVYLVNVVGNIGMIILITTDTQLHTPMYFFLCNLSFVDLGYSSAIAPRMLADFLTNHKVISFSSCATQFAFFVGFVDAECYVLAAMAYGRFVAICRPLHYSTFMSKQVCLALMLGSYLAGLVSLVAHTTLTFSLSYCGSNIINHFFCEIPPLLALSCSDTYISEILLFSLCGFIEFSTILIIFISYTFILVAIIRMRSAEGRLKAFSTCGSHLTGITLFYGTVMFMYLRPTSSYSLDQDKWASVFYTVIIPMLNPLIYSLRNKDVKAAFKKLIGKKSQ | |||||||||||||||||||||||||
| 1 | 3emlA | 0.15 | 0.17 | 0.89 | 3.60 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 2 | 5tgzA | 0.20 | 0.23 | 0.88 | 2.06 | Download | -RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL------------------------PLLGWNCEKLQSVDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM------ | |||||||||||||||||||
| 3 | 4iaqA | 0.15 | 0.22 | 0.87 | 2.16 | Download | ------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR----WVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL-------PPFFWRQAS--------------ECVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIIQLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK-- | |||||||||||||||||||
| 4 | 4djh | 0.15 | 0.23 | 0.91 | 1.53 | Download | -------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--- | |||||||||||||||||||
| 5 | 4yay | 0.16 | 0.24 | 0.91 | 1.25 | Download | KTTR-NAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHR-NVFF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL----- | |||||||||||||||||||
| 6 | 3emlA | 0.15 | 0.17 | 0.89 | 3.74 | Download | ---------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 7 | 4iaq | 0.19 | 0.21 | 0.85 | 1.72 | Download | ---------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQASECVVN--------------------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPI--HL-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK-- | |||||||||||||||||||
| 8 | 2z73A | 0.19 | 0.23 | 0.97 | 2.96 | Download | -ETYNPSIVVHPHWREFDQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDY---------ISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLT | |||||||||||||||||||
| 9 | 3emlA | 0.15 | 0.17 | 0.89 | 5.21 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 10 | 2ydoA | 0.14 | 0.16 | 0.92 | 6.00 | Download | -----------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| ||||||||||||||||||||||||||
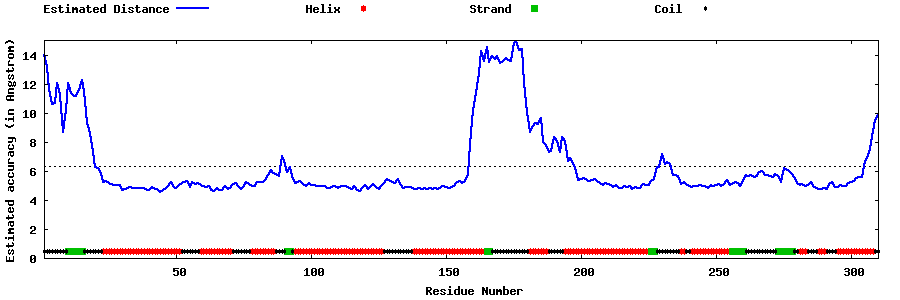
| Generated 3D models | Estimated local accuracy of models | ||
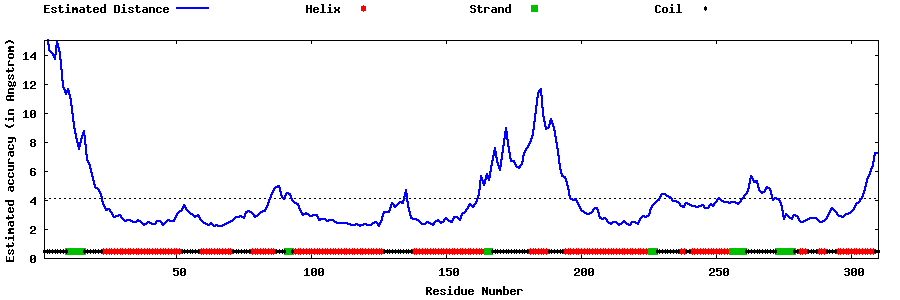 | |||
|
|
|||
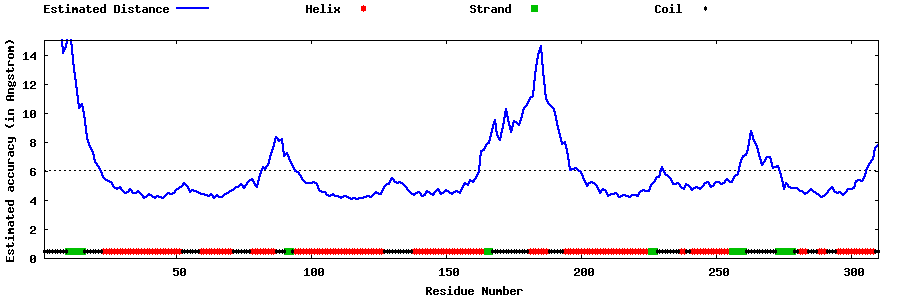 | |||
|
| |||
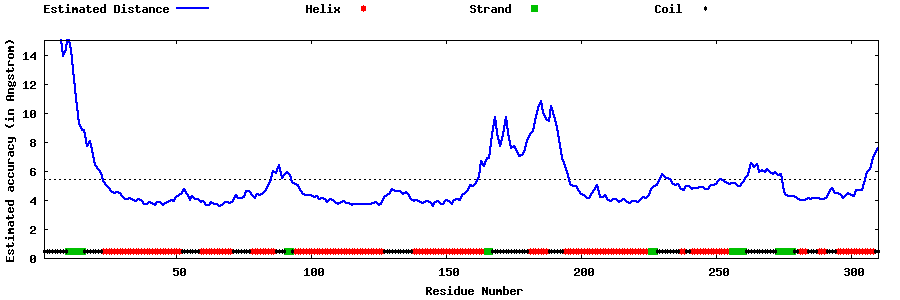 | |||
|
| |||
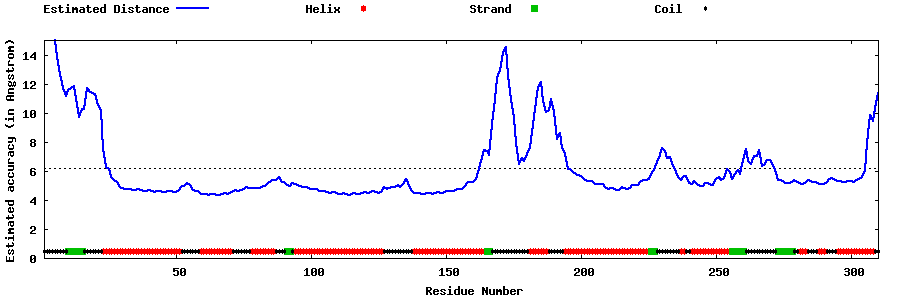 | |||
|
| |||
 | |||
|
| |||