

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MEWENHTILVEFFLKGLSGHPRLELLFFVLIFIMYVVILLGNGTLILISILDPHLHTPMYFFLGNLSFLDICYTTTSIPSTLVSFLSERKTISLSGCAVQMFLGLAMGTTECVLLGMMAFDRYVAICNPLRYPIIMSKDAYVPMAAGSWIIGAVNSAVQSVFVVQLPFCRNNIINHFTCEILAVMKLACADISDNEFIMLVATTLFILTPLLLIIVSYTLIIVSIFKISSSEGRSKASSTCSAHLTVVIIFYGTILFMYMKPKSKETLNSDDLDATDKIISMFYGVMTPMMNPLIYSLRNKDVKEAVKHLLNRRFFSK | |
| CCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCCCCCCSSSSSCCCHHHCCCCHHHCCCCHHHHHHHHHHHHHCCCCC | |
| 999888413677897689984369999999999999999988999798864888677578886779998786653578999998577996784899999999999999999999999986506650655448840388799999999999999999999999846078998955876358088888851576099999999999999998999999999999999338662546415436687899799998547631617898464432224667958998620725221136431464999999999986401589 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MEWENHTILVEFFLKGLSGHPRLELLFFVLIFIMYVVILLGNGTLILISILDPHLHTPMYFFLGNLSFLDICYTTTSIPSTLVSFLSERKTISLSGCAVQMFLGLAMGTTECVLLGMMAFDRYVAICNPLRYPIIMSKDAYVPMAAGSWIIGAVNSAVQSVFVVQLPFCRNNIINHFTCEILAVMKLACADISDNEFIMLVATTLFILTPLLLIIVSYTLIIVSIFKISSSEGRSKASSTCSAHLTVVIIFYGTILFMYMKPKSKETLNSDDLDATDKIISMFYGVMTPMMNPLIYSLRNKDVKEAVKHLLNRRFFSK | |
| 874623020210000000332401110013023312312333320120010013000000110200000001000031030001011652201010012011201200210030001001000000021020000003300000011001100210200000001010035130000000320001000210310022023312331331221023012200100010313622310110000000000011200100001032532344444533100000210332033003101010420130022003442238 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCCCCCCSSSSSCCCHHHCCCCHHHCCCCHHHHHHHHHHHHHCCCCC MEWENHTILVEFFLKGLSGHPRLELLFFVLIFIMYVVILLGNGTLILISILDPHLHTPMYFFLGNLSFLDICYTTTSIPSTLVSFLSERKTISLSGCAVQMFLGLAMGTTECVLLGMMAFDRYVAICNPLRYPIIMSKDAYVPMAAGSWIIGAVNSAVQSVFVVQLPFCRNNIINHFTCEILAVMKLACADISDNEFIMLVATTLFILTPLLLIIVSYTLIIVSIFKISSSEGRSKASSTCSAHLTVVIIFYGTILFMYMKPKSKETLNSDDLDATDKIISMFYGVMTPMMNPLIYSLRNKDVKEAVKHLLNRRFFSK | |||||||||||||||||||||||||
| 1 | 3emlA | 0.16 | 0.20 | 0.87 | 3.46 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISTG--FCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLW------LMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| 2 | 5tgzA | 0.22 | 0.24 | 0.87 | 2.13 | Download | -RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL------------------------PLLGWNCEKLQSVDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKT---VFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSM-------- | |||||||||||||||||||
| 3 | 5tgzA | 0.19 | 0.24 | 0.87 | 2.17 | Download | ----------GGRGENFMDIPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKD-SRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG----WNCEKLIFPHID------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHA-PDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------- | |||||||||||||||||||
| 4 | 4djh | 0.16 | 0.22 | 0.88 | 1.53 | Download | -------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS---A---ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----- | |||||||||||||||||||
| 5 | 4yay | 0.18 | 0.23 | 0.91 | 1.24 | Download | KTTRN-AYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIRDCRIADIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL------- | |||||||||||||||||||
| 6 | 3emlA | 0.17 | 0.20 | 0.87 | 3.73 | Download | ---------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAP------LWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| 7 | 3uon | 0.18 | 0.23 | 0.61 | 1.70 | Download | ------------------------VFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIV--GVRTVEDGECYIQF---------FSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSR------------------------------------------------------------------------------------------ | |||||||||||||||||||
| 8 | 2z73A | 0.19 | 0.22 | 0.94 | 2.67 | Download | -EWYNPSIVVHPHWREFDQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGAYTLE---GVLCNCSFDYI--------SRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLE------WVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCC | |||||||||||||||||||
| 9 | 3emlA | 0.17 | 0.20 | 0.87 | 4.66 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLM------YLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| 10 | 2ydoA | 0.15 | 0.16 | 0.90 | 5.54 | Download | -----------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLPFFCPDCSHA------PLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| ||||||||||||||||||||||||||
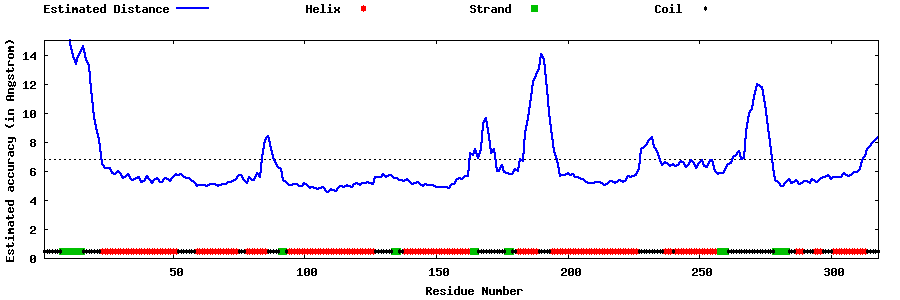
| Generated 3D models | Estimated local accuracy of models | ||
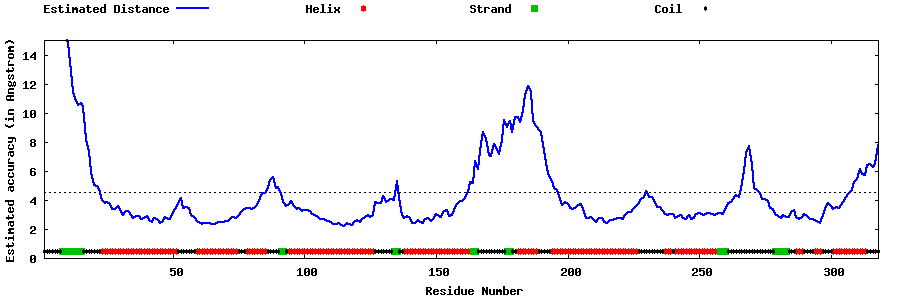 | |||
|
|
|||
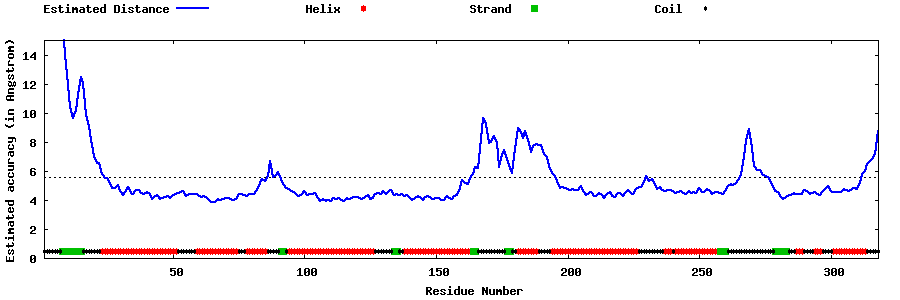 | |||
|
| |||
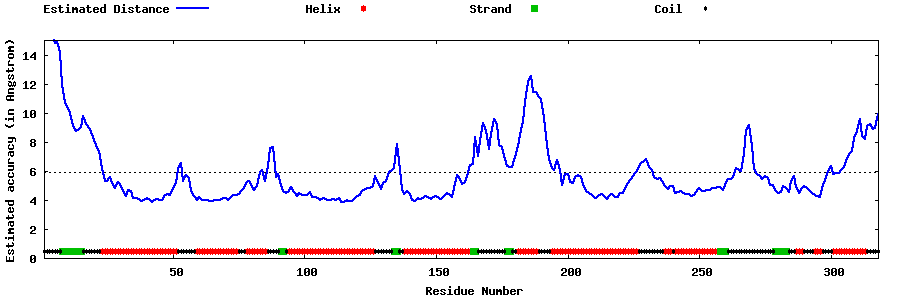 | |||
|
| |||
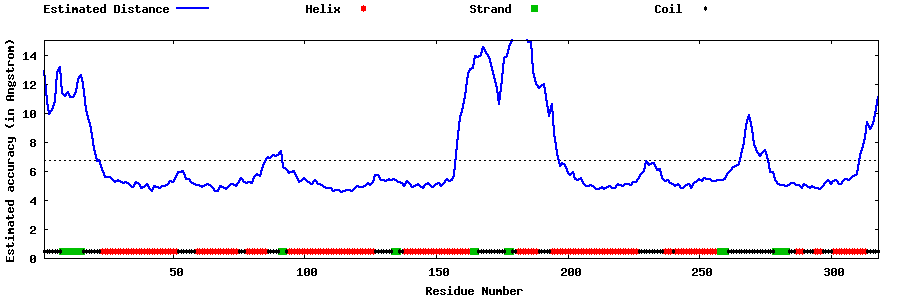 | |||
|
| |||
 | |||
|
| |||