

| 20 40 60 80 100 120 140 160 180 200 220 240 260 | |
| | | | | | | | | | | | | | | |
| MLGNYSSATEFFLLGFPGSQEVCRILFATFFLLYAVTVMGNVVIIITVCVDKCLQSPIYFFLGHLCVLEILITSTAVPFMLWGLLLPSTQIMSLTACAAQLYLYLSLGTLELALMGVMAVDRYVAVCNPLRYNIIMNSSTFIWVIIVSWVLGFLSEIWPVYATFQLTFCKSSVLDHFYCDRGQLLKVSCEDTLFREFILFLMAVFIIIGSLIPTIVSYTYIISTNLKIPSASGWRKSFSTCASHFTYVVIGYGSCLFLYVKPK | |
| CCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHHCCCCCSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHSSSSSCCC | |
| 98886420478885799884369999999999999999988999899972887677388885789999999984068999999762699777489999999999999999999999997651686275545883367758999999999999999999999972668889897788424858888783468639999999999999999899999999999999920865223751432548568848885420338982789 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 | |
| | | | | | | | | | | | | | | |
| MLGNYSSATEFFLLGFPGSQEVCRILFATFFLLYAVTVMGNVVIIITVCVDKCLQSPIYFFLGHLCVLEILITSTAVPFMLWGLLLPSTQIMSLTACAAQLYLYLSLGTLELALMGVMAVDRYVAVCNPLRYNIIMNSSTFIWVIIVSWVLGFLSEIWPVYATFQLTFCKSSVLDHFYCDRGQLLKVSCEDTLFREFILFLMAVFIIIGSLIPTIVSYTYIISTNLKIPSASGWRKSFSTCASHFTYVVIGYGSCLFLYVKPK | |
| 73424030110000000433401010013033313323333320020020033021000110300011002010010032001002454310001001001100120021003000200100000002212000200330010000101320233032201100304014323000000132100300022031103312331333333133203302320020001032463232010001002000202320010102338 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHHCCCCCSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHSSSSSCCC MLGNYSSATEFFLLGFPGSQEVCRILFATFFLLYAVTVMGNVVIIITVCVDKCLQSPIYFFLGHLCVLEILITSTAVPFMLWGLLLPSTQIMSLTACAAQLYLYLSLGTLELALMGVMAVDRYVAVCNPLRYNIIMNSSTFIWVIIVSWVLGFLSEIWPVYATFQLTFCKSSVLDHFYCDRGQLLKVSCEDTLFREFILFLMAVFIIIGSLIPTIVSYTYIISTNLKIPSASGWRKSFSTCASHFTYVVIGYGSCLFLYVKPK | |||||||||||||||||||||||||
| 1 | 3emlA | 0.17 | 0.25 | 0.87 | 2.91 | Download | -------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS----GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIIN-CFTFFCPD | |||||||||||||||||||
| 2 | 5tgzA | 0.15 | 0.21 | 0.87 | 2.01 | Download | --ENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKD--SRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-------GWNCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICW----------- | |||||||||||||||||||
| 3 | 5tgzA | 0.16 | 0.21 | 0.87 | 2.11 | Download | ---------GGRGENFMDIPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK--DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG----WNCEKLIFPHID------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHA-PDQARMDIELAKTLVLILVVLIICWGPLLAIMV | |||||||||||||||||||
| 4 | 3uon | 0.17 | 0.21 | 0.88 | 1.50 | Download | -------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVI-GYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIV--GVRTVEDGECYIQFF---------SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINISREKKVTRTILAILLAFIITWAPYNVMVLIPC | |||||||||||||||||||
| 5 | 4yay | 0.11 | 0.18 | 0.92 | 1.27 | Download | KTTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAM-EYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDD | |||||||||||||||||||
| 6 | 3emlA | 0.17 | 0.25 | 0.87 | 3.06 | Download | --------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISTG---FCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPD | |||||||||||||||||||
| 7 | 4iaq | 0.18 | 0.20 | 0.71 | 1.68 | Download | --------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTG-RWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQASE----------CVVN----------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSR----------------------------------- | |||||||||||||||||||
| 8 | 2rh1A | 0.16 | 0.19 | 0.85 | 1.97 | Download | ------------------DEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTF-GNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQM----HWYRATHQEAINCYAEE----TCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLNIFEMLRIDEGL-------------RLKIYKDTE | |||||||||||||||||||
| 9 | 3emlA | 0.17 | 0.25 | 0.87 | 3.84 | Download | -------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS----GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPD | |||||||||||||||||||
| 10 | 2ydoA | 0.18 | 0.25 | 0.90 | 4.38 | Download | ----------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA---ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWFTFFCPD | |||||||||||||||||||
| ||||||||||||||||||||||||||
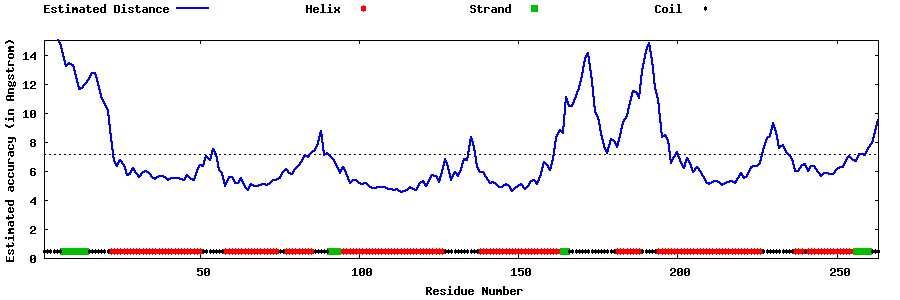
| Generated 3D models | Estimated local accuracy of models | ||
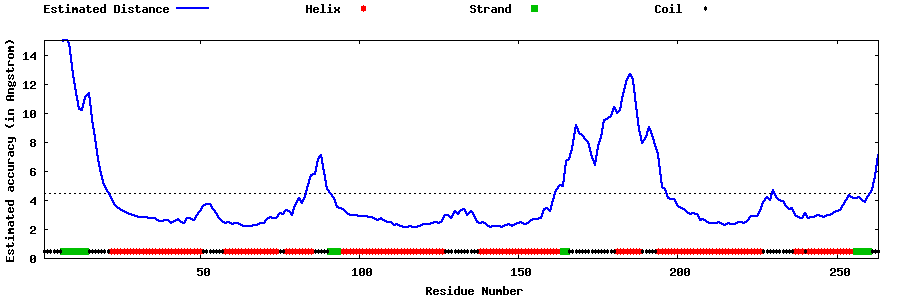 | |||
|
|
|||
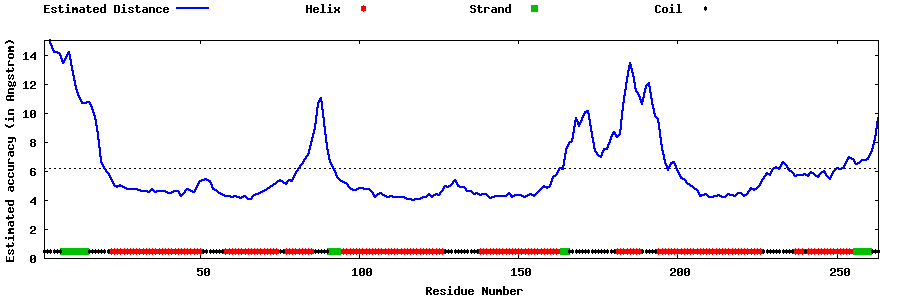 | |||
|
| |||
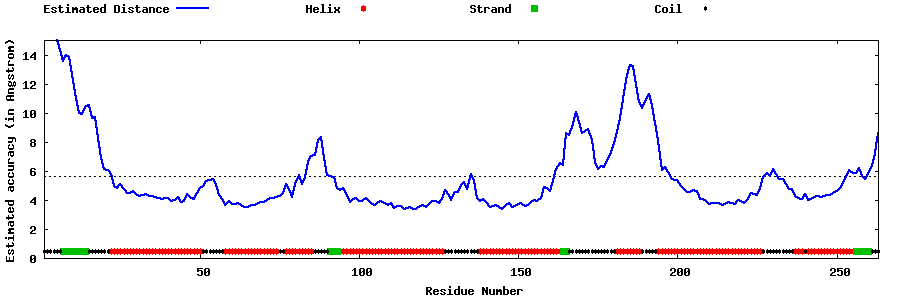 | |||
|
| |||
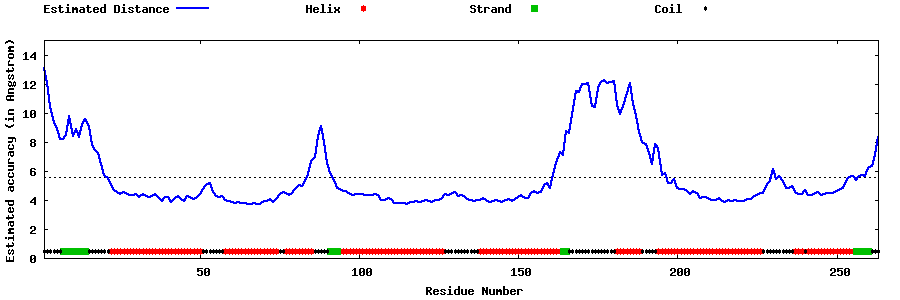 | |||
|
| |||
 | |||
|
| |||