

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | |
| | | | | | | | | | | | | | | | | | | | | | | |
| MEDLFSPSILPPAPNISVPILLGWGLNLTLGQGAPASGPPSRRVRLVFLGVILVVAVAGNTTVLCRLCGGGGPWAGPKRRKMDFLLVQLALADLYACGGTALSQLAWELLGEPRAATGDLACRFLQLLQASGRGASAHLVVLIALERRRAVRLPHGRPLPARALAALGWLLALLLALPPAFVVRGDSPSPLPPPPPPTSLQPGAPPAARAWPGERRCHGIFAPLPRWHLQVYAFYEAVAGFVAPVTVLGVACGHLLSVWWRHRPQAPAAAAPWSASPGRAPAPSALPRAKVQSLKMSLLLALLFVGCELPYFAARLAAAWSSGPAGDWEGEGLSAALRVVAMANSALNPFVYLFFQAGDCRLRRQLRKRLGSLCCAPQGGAEDEEGPRGHQALYRQRWPHPHYHHARREPLDEGGLRPPPPRPRPLPCSCESAF | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCCCCCCSSSSSCCCCCCSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 99888888799998677887667887688887888777478999999999998893688774440245347766666677357898999999999999965999999997378766842764479999999999999999999999899998589884999999999999999999999999713311456554442001467740123258754010136764078999999999999999999999999999999998706876521010123432112200788888888766679999999996599999999996656443226899999999999999878899987128673999999999998735887788666778778887767678988856365778878888899999999997656679 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | |
| | | | | | | | | | | | | | | | | | | | | | | |
| MEDLFSPSILPPAPNISVPILLGWGLNLTLGQGAPASGPPSRRVRLVFLGVILVVAVAGNTTVLCRLCGGGGPWAGPKRRKMDFLLVQLALADLYACGGTALSQLAWELLGEPRAATGDLACRFLQLLQASGRGASAHLVVLIALERRRAVRLPHGRPLPARALAALGWLLALLLALPPAFVVRGDSPSPLPPPPPPTSLQPGAPPAARAWPGERRCHGIFAPLPRWHLQVYAFYEAVAGFVAPVTVLGVACGHLLSVWWRHRPQAPAAAAPWSASPGRAPAPSALPRAKVQSLKMSLLLALLFVGCELPYFAARLAAAWSSGPAGDWEGEGLSAALRVVAMANSALNPFVYLFFQAGDCRLRRQLRKRLGSLCCAPQGGAEDEEGPRGHQALYRQRWPHPHYHHARREPLDEGGLRPPPPRPRPLPCSCESAF | |
| 83422313211221000123333212011333343332220010000011101200020010000000023443433220000000000020000000000213000001232201010000100300100000000000000001012000101233310100000000000000010000000133332322222222333221011312332101131232430010000001122133211201020001000101332443534544453443334444323332200000000000000000100000000000133233520210000000000000000000000000430340033025102200023444465453433333333333333343433434345433353263445221335348 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCCCCCCSSSSSCCCCCCSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MEDLFSPSILPPAPNISVPILLGWGLNLTLGQGAPASGPPSRRVRLVFLGVILVVAVAGNTTVLCRLCGGGGPWAGPKRRKMDFLLVQLALADLYACGGTALSQLAWELLGEPRAATGDLACRFLQLLQASGRGASAHLVVLIALERRRAVRLPHGRPLPARALAALGWLLALLLALPPAFVVRGDSPSPLPPPPPPTSLQPGAPPAARAWPGERRCHGIFAPLPRWHLQVYAFYEAVAGFVAPVTVLGVACGHLLSVWWRHRPQAPAAAAPWSASPGRAPAPSALPRAKVQSLKMSLLLALLFVGCELPYFAARLAAAWSSGPAGDWEGEGLSAALRVVAMANSALNPFVYLFFQAGDCRLRRQLRKRLGSLCCAPQGGAEDEEGPRGHQALYRQRWPHPHYHHARREPLDEGGLRPPPPRPRPLPCSCESAF | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.19 | 0.17 | 0.68 | 2.45 | Download | -----------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYT-----KMKTATNIYIFNLALADALATSTLPFQSAKYLM----TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-------------DGAVVCMLQFPS----------PSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLL---------------SGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLD---ENFKRCFRQLCRKPCG----------------------------------------------------------- | |||||||||||||||||||
| 2 | 5gliA | 0.16 | 0.20 | 0.74 | 3.85 | Download | --------------------------SPPPCQGPIEIKETFKYINTVVSCLVFVLGIIGNSTLLYIIYKNK-----CMRNGPNILIASLALGDLLHIVIAIPINVYKLLA--EDWPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWSRVPKWTAVEIVLIWVVSVVLAVPEAIGFDIITMDYKGSYLR---------ICLLHPVQKTAF-------MQFYATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLRDEGGGSGGDEAEKRTGTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQNDPSFLLVLDYIGINMASLNSCANPIALYLVS---KRFKNAFKSALCC-------------------------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.20 | 0.17 | 0.68 | 3.06 | Download | -----------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYT-----KMKTATNIYIFNLALADALATST-LPFQSAKYLMET--WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-------------GAVVCMLQFPSP----------SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSK---------------EKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDE---NFKRCFRQLCRKPCG----------------------------------------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.14 | 0.17 | 0.83 | 1.54 | Download | ----------------DLRDNETWWYNPSIIVHFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKT-----KSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFL-KKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEG----------------------VLCNCSFDYISRDS-TTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRL--NAKELRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLE---WVTPYAAQLPVMFAKASAIHNPMIYSVSHP---KFREAISQTFPWVLTCCQFDDKETEDDK-DAETEI-----P-------------AGESSDAAPSADAAQMKE-- | |||||||||||||||||||
| 5 | 4rnb | 0.19 | 0.21 | 0.70 | 1.16 | Download | -------------------------------------PKEYEWVLIAGYIIVFVVALIGNVLVCVAVWKNH-----HMRTVTNYFIVNLSLADVLVTITCLPATLVVDITE--TWFFGQSLCKVIPYLQTVSVSVSVLTLSCIALDRWYAICHPST-AKRARNSIVIIWIVSCIIMIPQAIVMECSTVFK----------TTLFTVCDERWGGE------------IYPKMYHICFFLVTYMAPLCLMVLAYLQIFRKLWCRQGIDCSFWNESRGQRSDLMSFSKQIRARRKTARMLMVVLLVFAICYLPISILNVLKRVFGMFHDRETVYAWFTFSHWLVYANSAANPIIYNFLSG---KFREEFKAAFSC-------------------------------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.20 | 0.17 | 0.68 | 2.66 | Download | -------------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY-----TKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMET--WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-------------GAVVCMLQFPS----------PSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSG---------------SKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLD---ENFKRCFRQLCRKPCG----------------------------------------------------------- | |||||||||||||||||||
| 7 | 3uon | 0.17 | 0.19 | 0.66 | 1.74 | Download | -----------------------------------------VVFIVLVAGSLSLVTIIGNILVMVSIKVNR-----HLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVI--GYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKIAAAWVLSFILWAPAILFWQFIVGVRTVE----------DGE----------CYIQFFSN-----AAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEEGGHAARNTKTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPC----IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKH--------------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.21 | 0.17 | 0.68 | 3.80 | Download | --------------------------------GSPGSSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY-----TKMKTATNIYIFNLALADALATST-LPFQSAKYLME--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----------------------GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSG---------------SKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLD-------ENFKRCFRQLCRKPCG------------------------------------------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.19 | 0.17 | 0.68 | 2.53 | Download | -----------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYT-----KMKTATNIYIFNLALADALATSTLPFQSAKYLM----TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-------------GAVVCMLQFPS----------PSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---------------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLD---ENFKRCFRQLCRKPCG----------------------------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.20 | 0.15 | 0.67 | 2.27 | Download | -------------------------GSHSLPQTGSPSM-VTAITIMALYSIVCVVGLFGNFLVMYVIVRYT-----KMKTATNIYIFNLALADALAT-STLPFQSVNYLMGT--WPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYR-----------------------QGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLK---------------SVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPETT-FQTVSWHFCIALGYTNSCLNPVLYAFLD---ENFKRCF-------------------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
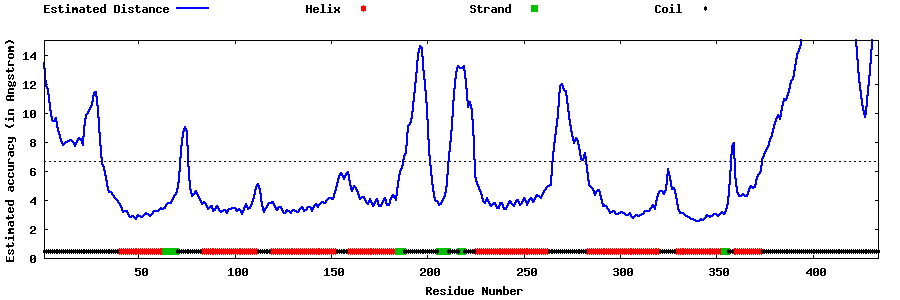 | |||
|
|
|||
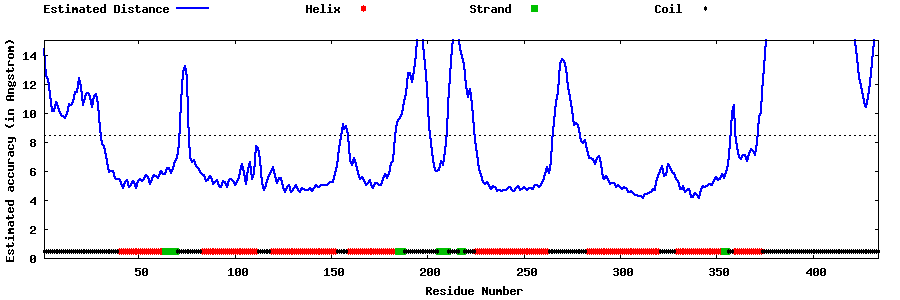 | |||
|
| |||
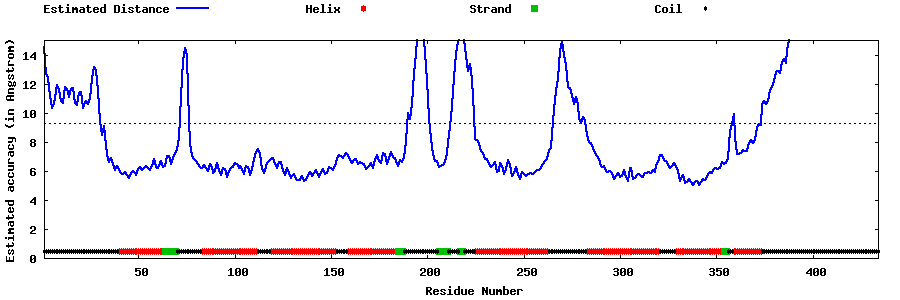 | |||
|
| |||
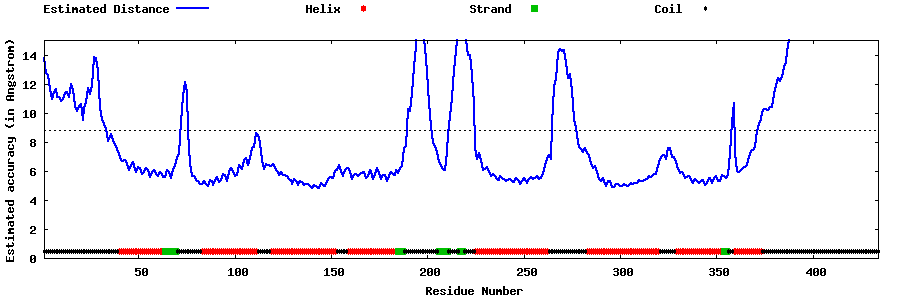 | |||
|
| |||
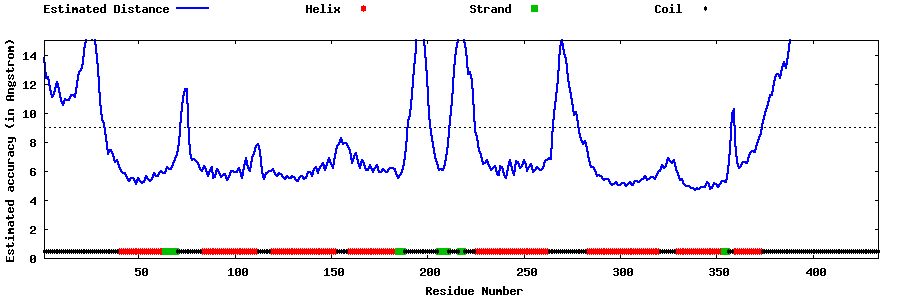 | |||
|
| |||