

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MDIPQNITEFFMLGLSQNSEVQRVLFVVFLLIYVVTVCGNMLIVVTITSSPTLASPVYFFLANLSFIDTFYSSSMAPKLIADSLYEGRTISYECCMAQLFGAHFLGGVEIILLTVMAYDRYVAICKPLHNTTIMTRHLCAMLVGVAWLGGFLHSLVQLLLVLWLPFCGPNVINHFACDLYPLLEVACTNTYVIGLLVVANSGLICLLNFLMLAASYIVILYSLRSHSADGRCKALSTCGAHFIVVALFFVPCIFTYVHPFSTLPIDKNMALFYGILTPMLNPLIYTLRNEEVKNAMRKLFTW | |
| CCCCCCSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCSSCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHCSSSSSCCCCCCCCCHHSSHHHHHHHHHHCCCHHHCCCCHHHHHHHHHHHCC | |
| 99997300578816999821589999999999999998788745523307999997899987599876512234719999997439967848999999999999999899999999965288746220260026774899999999999999999999995228998988367764284897878203414445468776679999999999999999999940257302229998998799934256306325886889999730213256652244000374335619999999988369 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MDIPQNITEFFMLGLSQNSEVQRVLFVVFLLIYVVTVCGNMLIVVTITSSPTLASPVYFFLANLSFIDTFYSSSMAPKLIADSLYEGRTISYECCMAQLFGAHFLGGVEIILLTVMAYDRYVAICKPLHNTTIMTRHLCAMLVGVAWLGGFLHSLVQLLLVLWLPFCGPNVINHFACDLYPLLEVACTNTYVIGLLVVANSGLICLLNFLMLAASYIVILYSLRSHSADGRCKALSTCGAHFIVVALFFVPCIFTYVHPFSTLPIDKNMALFYGILTPMLNPLIYTLRNEEVKNAMRKLFTW | |
| 87562200000000004244001000131333233233323300000200430400001002300120011000000200000035321000500010001103311310200020030000000221101000033100000220131023013001310230300011220001002220030001202300030023013202310331331121002001321461222011003002100301321010000003331330000003102310330030000205402300331167 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCSSCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHCSSSSSCCCCCCCCCHHSSHHHHHHHHHHCCCHHHCCCCHHHHHHHHHHHCC MDIPQNITEFFMLGLSQNSEVQRVLFVVFLLIYVVTVCGNMLIVVTITSSPTLASPVYFFLANLSFIDTFYSSSMAPKLIADSLYEGRTISYECCMAQLFGAHFLGGVEIILLTVMAYDRYVAICKPLHNTTIMTRHLCAMLVGVAWLGGFLHSLVQLLLVLWLPFCGPNVINHFACDLYPLLEVACTNTYVIGLLVVANSGLICLLNFLMLAASYIVILYSLRSHSADGRCKALSTCGAHFIVVALFFVPCIFTYVHPFSTLPIDKNMALFYGILTPMLNPLIYTLRNEEVKNAMRKLFTW | |||||||||||||||||||||||||
| 1 | 3emlA | 0.21 | 0.25 | 0.89 | 3.37 | Download | ------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVPMNYMVYFN-----------FFACVLVPLLLMLGVYLRIFLAARQLVHAAKSLAIIVGLFALCWLPLH-IINCFTFFCPDCSHAWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRS | |||||||||||||||||||
| 2 | 5tgzA | 0.22 | 0.26 | 0.91 | 2.27 | Download | -ENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL--------PLLGWNCEK---------LQSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARELAKTLVLILVVLIICWGPLLAIMVKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSM--- | |||||||||||||||||||
| 3 | 5tgzA | 0.19 | 0.26 | 0.91 | 2.08 | Download | -GGRGENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFH-VFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLCEKLQSVCS-----------------------DIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHRMDIELAKTLVLILVVLIICWGPLLAIMVGKMNLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF-- | |||||||||||||||||||
| 4 | 4djh | 0.16 | 0.25 | 0.92 | 1.53 | Download | -----------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDDD---YSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSLSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP | |||||||||||||||||||
| 5 | 4yay | 0.19 | 0.26 | 0.93 | 1.21 | Download | KTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK-----KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL-- | |||||||||||||||||||
| 6 | 5tgzA | 0.23 | 0.26 | 0.92 | 3.50 | Download | -ENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAV--------LPLLGWNCEK--------LQSVCSDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQAIELAKTLVLILVVLIICWGPLLAIMVYDVFGIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF-- | |||||||||||||||||||
| 7 | 4iaq | 0.24 | 0.26 | 0.86 | 1.71 | Download | ------------------LPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQASE----------CVVNT-------D---HILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADRERKATKTLGIILGAFIVCWLPFFIILVMPIH--LAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIR- | |||||||||||||||||||
| 8 | 4buoA | 0.17 | 0.23 | 0.90 | 3.07 | Download | ---NSD------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGCTPIV---------DTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQAVVIAFVVCWLPYHVRRLMFCYI-----------SDEQMLTNALVYVSAAINPILYNLVSANFRQVFLSTL-- | |||||||||||||||||||
| 9 | 5tgzA | 0.23 | 0.26 | 0.91 | 4.65 | Download | GENFMDIECFMVL----NPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVF-HRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG---------NCEK---------LQSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMN-FAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF-- | |||||||||||||||||||
| 10 | 2ydoA | 0.19 | 0.23 | 0.92 | 5.35 | Download | ---------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRLKQMESTLQKEVHAAKSLAIIVGLFALCCFTFFCPDCSHAPLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRS | |||||||||||||||||||
| ||||||||||||||||||||||||||
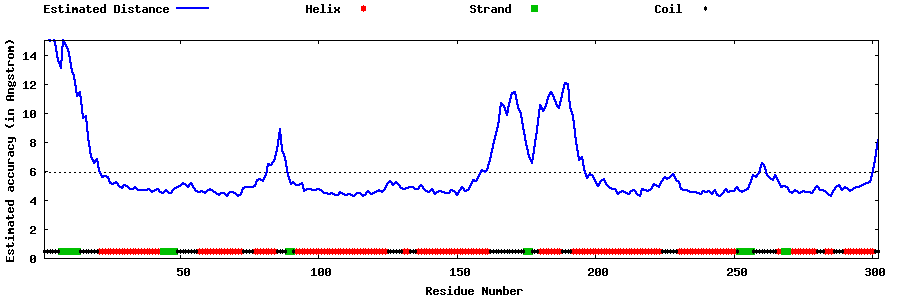
| Generated 3D models | Estimated local accuracy of models | ||
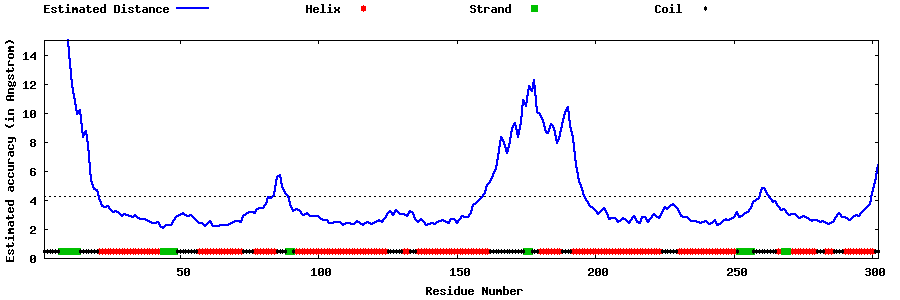 | |||
|
|
|||
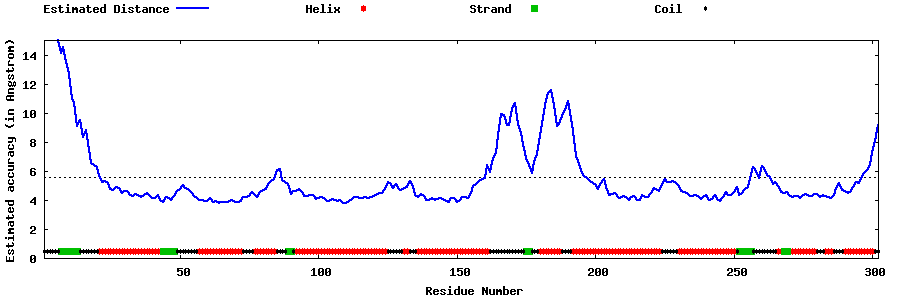 | |||
|
| |||
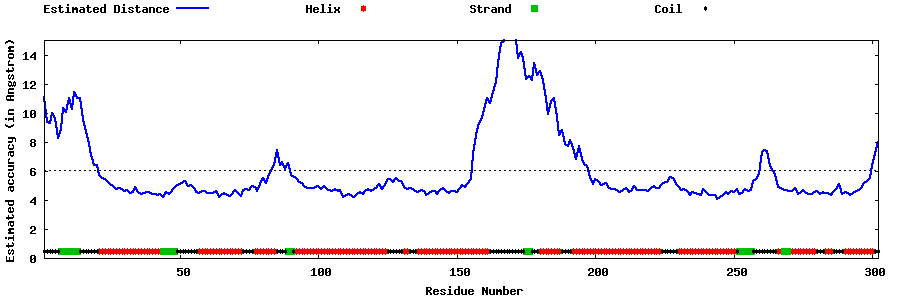 | |||
|
| |||
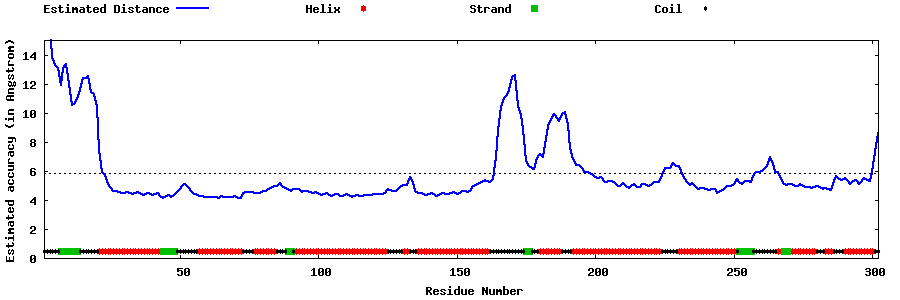 | |||
|
| |||
 | |||
|
| |||