

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MERQNQSCVVEFILLGFSNYPELQGQLFVAFLVIYLVTLIGNAIIIVIVSLDQSLHVPMYLFLLNLSVVDLSFSAVIMPEMLVVLSTEKTTISFGGCFAQMYFILLFGGAECFLLGAMAYDRFAAICHPLNYQMIMNKGVFMKLIIFSWALGFMLGTVQTSWVSSFPFCGLNEINHISCETPAVLELACADTFLFEIYAFTGTFLIILVPFLLILLSYIRVLFAILKMPSTTGRQKAFSTCAAHLTSVTLFYGTASMTYLQPKSGYSPETKKVMSLSYSLLTPLLNLLIYSLRNSEMKRALMKLWRRRVVLHTI | |
| CCCCCCCSSSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHCCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCSSSSSSHHCHHHCCCCHHHCCCCHHHHHHHHHHHHHHHHHCCC | |
| 99988731324234469998517999999999999999998899989997388755758999888999987777028899989855799678599999999999999999999999998650664065545883158879999999999999999999999990417899895788645818889885457519999999999999999799999999999999822867366663202758689979998721541781789999887781898745254532024533146599999999999761101269 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MERQNQSCVVEFILLGFSNYPELQGQLFVAFLVIYLVTLIGNAIIIVIVSLDQSLHVPMYLFLLNLSVVDLSFSAVIMPEMLVVLSTEKTTISFGGCFAQMYFILLFGGAECFLLGAMAYDRFAAICHPLNYQMIMNKGVFMKLIIFSWALGFMLGTVQTSWVSSFPFCGLNEINHISCETPAVLELACADTFLFEIYAFTGTFLIILVPFLLILLSYIRVLFAILKMPSTTGRQKAFSTCAAHLTSVTLFYGTASMTYLQPKSGYSPETKKVMSLSYSLLTPLLNLLIYSLRNSEMKRALMKLWRRRVVLHTI | |
| 87452302001000000033240111001301331331333332002001003300000011020000100101003203001101164320100002201200130021002000100200000002102000100330000000100110031031001000002003624010000032000200021031002202331333133123101301320000001031362231000000000000001120000000103163364241000021033213300300001042013002200233211443 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCSSSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHCCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCSSSSSSHHCHHHCCCCHHHCCCCHHHHHHHHHHHHHHHHHCCC MERQNQSCVVEFILLGFSNYPELQGQLFVAFLVIYLVTLIGNAIIIVIVSLDQSLHVPMYLFLLNLSVVDLSFSAVIMPEMLVVLSTEKTTISFGGCFAQMYFILLFGGAECFLLGAMAYDRFAAICHPLNYQMIMNKGVFMKLIIFSWALGFMLGTVQTSWVSSFPFCGLNEINHISCETPAVLELACADTFLFEIYAFTGTFLIILVPFLLILLSYIRVLFAILKMPSTTGRQKAFSTCAAHLTSVTLFYGTASMTYLQPKSGYSPETKKVMSLSYSLLTPLLNLLIYSLRNSEMKRALMKLWRRRVVLHTI | |||||||||||||||||||||||||
| 1 | 3emlA | 0.21 | 0.23 | 0.89 | 3.55 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISTG--FCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINC-FTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-- | |||||||||||||||||||
| 2 | 5tgzA | 0.18 | 0.20 | 0.90 | 2.17 | Download | -RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-------GWNCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFSMLCLLNSTVNPIIYALRSKDLRHAFRSM---------- | |||||||||||||||||||
| 3 | 5tgzA | 0.18 | 0.20 | 0.87 | 2.22 | Download | ----------GGRGENFMDIPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKD-SRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG----WNCEKLIFPHID------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQADIELAKTLVLILVVLIICWGPLLAIMVDVFGKMNKLTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------- | |||||||||||||||||||
| 4 | 4djh | 0.14 | 0.23 | 0.89 | 1.54 | Download | -------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------- | |||||||||||||||||||
| 5 | 4yay | 0.20 | 0.23 | 0.90 | 1.24 | Download | KTTRN-AYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVF-F------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL--------- | |||||||||||||||||||
| 6 | 3emlA | 0.21 | 0.23 | 0.88 | 3.82 | Download | ---------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-- | |||||||||||||||||||
| 7 | 4iaq | 0.22 | 0.23 | 0.84 | 1.71 | Download | --------------------LPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFW-RQASECVVN----------T-------D---HILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH--LAIF-DFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------ | |||||||||||||||||||
| 8 | 2z73A | 0.18 | 0.21 | 0.95 | 2.98 | Download | -ETYNPSIVVHPHWREFDQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGTLEGVLCNDYISR-------------DSTTRSNILCMFILG---FFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQF | |||||||||||||||||||
| 9 | 3emlA | 0.20 | 0.23 | 0.89 | 4.77 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-- | |||||||||||||||||||
| 10 | 2ydoA | 0.18 | 0.22 | 0.92 | 5.66 | Download | -----------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQE | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
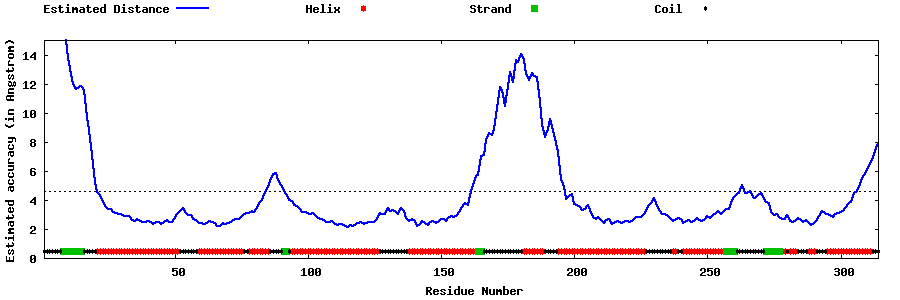 | |||
|
|
|||
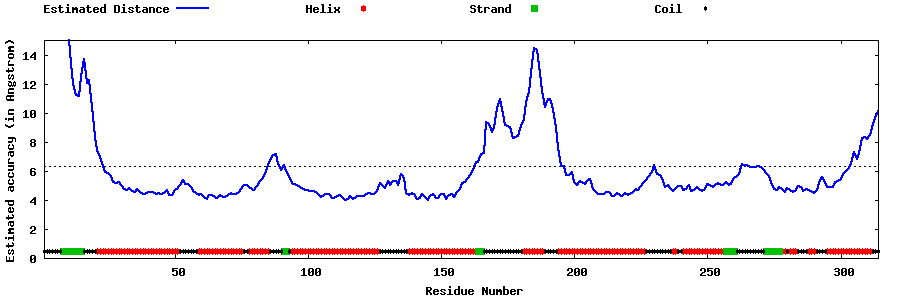 | |||
|
| |||
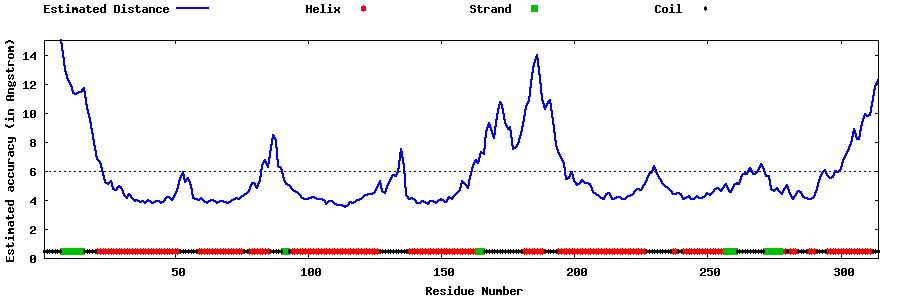 | |||
|
| |||
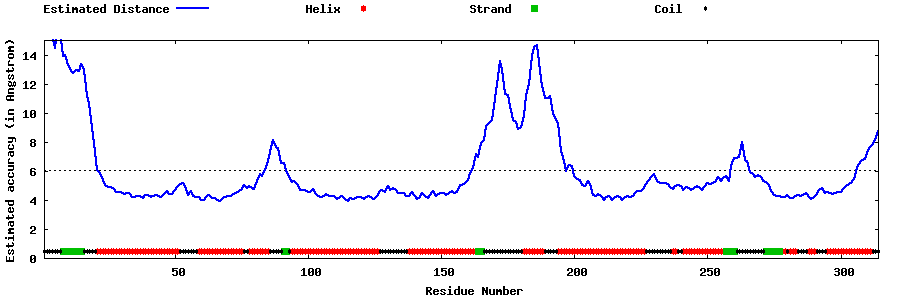 | |||
|
| |||
 | |||
|
| |||