

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MASKDFAIGMILLSQIMVGFLGNFFLLYHYSFLCFTRGMLQSTDLILKHLTIANSLVILSKGIPQTMAAFGLKDSLSDIGCKFVFYVHRVGRAVCVGNACLLSVFQVITISPSEFRWAELKLHAHKYIRSFILVLCWILNTLVNITVLLHVTGKWNSINSTKTNDYGYCSGGSRSRIPHSLHIVLLSSLDVLCLGLMTLASGSMVFILHRHKQQVQHIHGTNLSARSSPESRVTQSILVLVSTLCYFTRSPPSLHMSLFPNPSWWLLNTSALITACFPMVSPFVLMSRHPRIPRLGSACCGRNPQFPKLVR | |
| CCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSSSSSSSHHHCCCCHHHHHHHHHHHHSSSCCCCCHHHHHHHHCCCSSHHHHHHHHHHHHHHHHHCSSSSSSSCCCCCCCCCCCCCCSSSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHSSHHHHHHHHCHHHCHHHSSCCCCCHHHHHHHHHCCCCCCCCCCC | |
| 98788899999999999999999999999999998388889799999999999999999985899999975522489983289999807737203578999998761897189707665425347633799999999999987612579998277045555214675534316746789999999999989999999877889999999978988633089999998859999999999999999999878987566321798221009999960231536456013896278999999686788976569 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MASKDFAIGMILLSQIMVGFLGNFFLLYHYSFLCFTRGMLQSTDLILKHLTIANSLVILSKGIPQTMAAFGLKDSLSDIGCKFVFYVHRVGRAVCVGNACLLSVFQVITISPSEFRWAELKLHAHKYIRSFILVLCWILNTLVNITVLLHVTGKWNSINSTKTNDYGYCSGGSRSRIPHSLHIVLLSSLDVLCLGLMTLASGSMVFILHRHKQQVQHIHGTNLSARSSPESRVTQSILVLVSTLCYFTRSPPSLHMSLFPNPSWWLLNTSALITACFPMVSPFVLMSRHPRIPRLGSACCGRNPQFPKLVR | |
| 64333211111032123333333320132100000353322111000100131122103012311000000232114121000010101011110000000001200000013322023022301210101101230331131223000202033333233333331101012333321100010102312311000100100000001212320220234532433311320010000000111131113200100013342100100110201211200000000214201200200032335114327 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSSSSSSSHHHCCCCHHHHHHHHHHHHSSSCCCCCHHHHHHHHCCCSSHHHHHHHHHHHHHHHHHCSSSSSSSCCCCCCCCCCCCCCSSSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHSSHHHHHHHHCHHHCHHHSSCCCCCHHHHHHHHHCCCCCCCCCCC MASKDFAIGMILLSQIMVGFLGNFFLLYHYSFLCFTRGMLQSTDLILKHLTIANSLVILSKGIPQTMAAFGLKDSLSDIGCKFVFYVHRVGRAVCVGNACLLSVFQVITISPSEFRWAELKLHAHKYIRSFILVLCWILNTLVNITVLLHVTGKWNSINSTKTNDYGYCSGGSRSRIPHSLHIVLLSSLDVLCLGLMTLASGSMVFILHRHKQQVQHIHGTNLSARSSPESRVTQSILVLVSTLCYFTRSPPSLHMSLFPNPSWWLLNTSALITACFPMVSPFVLMSRHPRIPRLGSACCGRNPQFPKLVR | |||||||||||||||||||||||||
| 1 | 4grvA | 0.13 | 0.20 | 0.90 | 1.21 | Download | DIYSKVLVTAIYLALFVVGTVGNSVTLFTLAR-----KSLQTVHYHLGSLALSDLLILLLAMPVELYNFIWVHHAFGDAGCRGYYFLRDACTYATALNVASLSVARYLAIC----HPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNRSADGTHPGGL-VCTPIVDTATVKVVIQVNTFMSFLFPMLVISILNTVIANKLTVM-------------GSVQALRHGVLVARAVVIAFVVCWLPYHVRRLMFCYISDQWTTFLFDFYHYFYMLTNALAYASS--AINPILYNLVSAN------FR | |||||||||||||||||||
| 2 | 5tjvA | 0.12 | 0.21 | 0.86 | 1.92 | Download | --SQQLAIAVLSLTLGTFTVLENLLVL--CVILHSRSLRCRPSYHFIGSLAVADLLGSVIF-VYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRP----KAVVAFCLMWTIAIVIAVLPLL---------GWNCEKLQSVCSDIFPHIDETYLMFWIGVT--SVLLLFIVYAYMYILWKAGKRAMSFSD--------QARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFR-------------- | |||||||||||||||||||
| 3 | 4xt1A | 0.13 | 0.20 | 0.86 | 1.33 | Download | LNQSKPVTLFLYGVVFLFGSIGNFLVIFTITW----RRRIQSGDVYFINLAAADLLFVCT--LPLWMQYLLDSVP-----CTLLTACFYVAMFASLCFITEIALDRYYAIVYMRYR--------PVKQACLFSIFWWIFAVIIAIPHFMVV-----TKKDNQCMTDY---DYLEVSYPIILNVELMLGAFVIPLSVISYCYYRISRIVA----------VSQSRHKGRIVRVLIAVVLVFIIFWLPYHLTLFVDTLKLLKWISSRALILTESLAFCHCCLNPLLYVFVGTKFRQELHCLLAEFR------- | |||||||||||||||||||
| 4 | 4djh | 0.15 | 0.24 | 0.93 | 1.54 | Download | SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR---YTKMKTATNIYIFNLALADALVTTT-MPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPV-KALDF--RTPLKAKII-NICIWLLSSSVGISAIVLGGTKVR-----EDVDVIECSLQFPDDDYDLFMICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIEKTAYREKDRNLRRITRLVLVVVAVFVVCTPIHIILVEALGS-AALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------- | |||||||||||||||||||
| 5 | 3odu | 0.12 | 0.17 | 0.94 | 1.27 | Download | ANFNKIFLPTIYSIIFLTGIVGNGLVILVM----GYQKLRSMTDKYRLHLSVADLLFVIT--LPFWAVDAVANWYFGNFLCKAVHVIYTVNLYSSVWILAFISLDRYLAIVATNSQ-RPRKLLAEK----VVYVGVWIPALLLTIPDFIFANVSE-------ADDRYICDRFYPNDLWVVVFQFHIMVGLILPGIVILSCYCIIISKLSHSGSNIFEMLRIAYGSKGHQKRKALKTTVILILAFFACWLPYYIGISSFILLEVHKWISITEALAFFHCCLNPILYAFLGAKFKTSAQHALTSGRPLEVLFQ | |||||||||||||||||||
| 6 | 4grvA | 0.11 | 0.20 | 0.88 | 1.35 | Download | -IYSKVLVTAIYLALFVVGTVGNSVTLFTLAR----KSLQSTVHYHLGSLALSDLLILLLAMPVELYNFIWVHHAFGDAGCRGYYFLRDACTYATALNVASLSVARYLAIC----HPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNRSADGTH-PGGLVCTPIVDTATVKVVIQVNTFMSFLFPMLVISILNTVIANKLTVMS------------GSVQALRHGVLVARAVVIAFVVCWLPYHVRRLMFCYIFYHYFYMLTNALAYASSAINPILYNLVSANFRQV---------------- | |||||||||||||||||||
| 7 | 3uon | 0.12 | 0.19 | 0.91 | 1.71 | Download | ----VVFIVLVAGSLSLVTIIGNILVMVSI---KVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGV-R-TVEDG------ECYIQFFSN--AAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEGTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM---------- | |||||||||||||||||||
| 8 | 4n6hA | 0.15 | 0.21 | 0.91 | 2.02 | Download | SLALAIAITALYSAVCAVGLLGNVLVMF---GIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-------------GVCMLQFPSYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV------RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------ | |||||||||||||||||||
| 9 | 4djhA | 0.16 | 0.20 | 0.89 | 1.31 | Download | SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR----TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPF-GDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVHPVKALDFRTPLK-----AKIINICIWLLSSSVGISAIVLGGTK-----VREDVDVIECSLQFPDSWWDLFMKICVFIFAVIPVLIIIVCYTLMILRLKSVRLLSDR-----------NLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAASSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFC-----FP---- | |||||||||||||||||||
| 10 | 4ww3A | 0.14 | 0.19 | 0.95 | 1.74 | Download | PDAVYYSLGIFIGICGIIGCGGNGIVIYLFTK---TKSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFLKKWFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIRPMAASK-----KMSHRRAFIMIIFVWLWSVLWAIGPIF------GWGAYTLEGVLCNCSFISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNAKGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASPMIYSVSHPKFREAISQTFPWVLTCCQFDD | |||||||||||||||||||
| ||||||||||||||||||||||||||
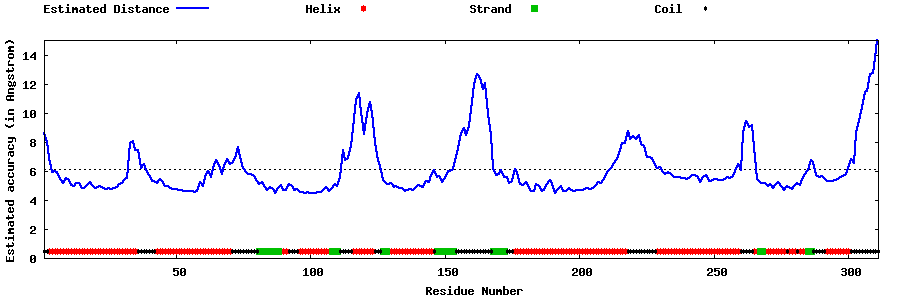
| Generated 3D models | Estimated local accuracy of models | ||
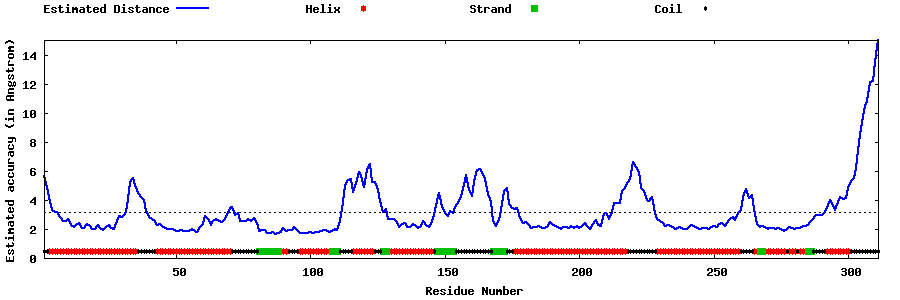 | |||
|
|
|||
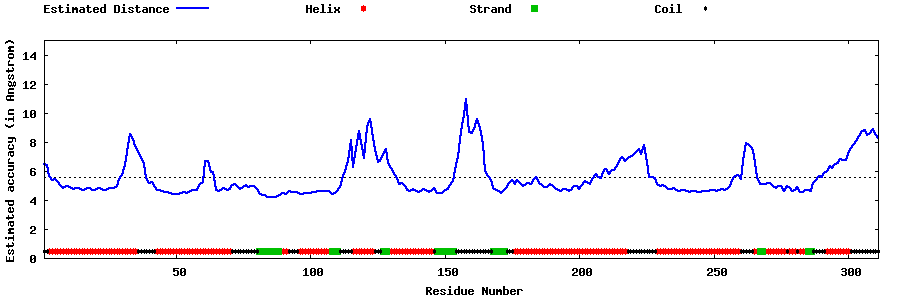 | |||
|
| |||
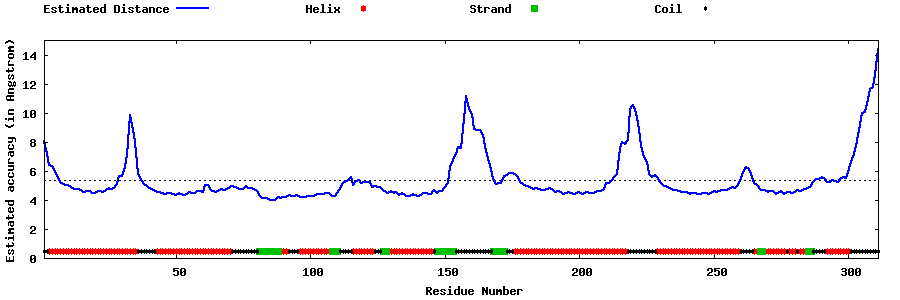 | |||
|
| |||
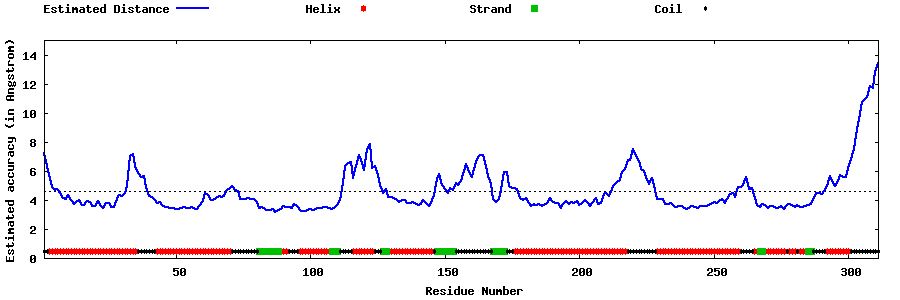 | |||
|
| |||
 | |||
|
| |||