

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MVGDTLKLLSPLMTRYFFLLFYSTDSSDLNENQHPLDFDEMAFGKVKSGISFLIQTGVGILGNSFLLCFYNLILFTGHKLRPTDLILSQLALANSMVLFFKGIPQTMAAFGLKYLLNDTGCKFVFYYHRVGTRVSLSTICLLNGFQAIKLNPSICRWMEIKIRSPRFIDFCCLLCWAPHVLMNASVLLLVNGPLNSKNSSAKNNYGYCSYKASKRFSSLHAVLYFSPDFMSLGFMVWASGSMVFFLYRHKQQVQHNHSNRLSCRPSQEARATHTIMVLVSSFFVFYSVHSFLTIWTTVVANPGQWIVTNSVLVASCFPARSPFVLIMSDTHISQFCFACRTRKTLFPNLVVMP | |
| CCCHHHHHHHHHHHHHHHHHSSCCCCCHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCSSSSSSSSSHHHCCCHHHHHHHHHHHHSSSSCCCCHHHHHHHHHCCCSSSHHHHHHHHHHHHHHCCSSSSSSCCCCCCCCCCCCCCSSSSSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHCHHHHHCCCCCHHHHHHHHHCCCCCCCCSSSCC | |
| 97514556325677456520325886007756766777877689999999999999999999999999999999817888968999999999999999998489999997463148987579999982572611278999998853099758954876645645867708889999999998556469997288660045321353721645558999999999999999999999999999999998678652533899999988799999999999999999998889999999991689810100999996421464857753589418899999818678999836179 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MVGDTLKLLSPLMTRYFFLLFYSTDSSDLNENQHPLDFDEMAFGKVKSGISFLIQTGVGILGNSFLLCFYNLILFTGHKLRPTDLILSQLALANSMVLFFKGIPQTMAAFGLKYLLNDTGCKFVFYYHRVGTRVSLSTICLLNGFQAIKLNPSICRWMEIKIRSPRFIDFCCLLCWAPHVLMNASVLLLVNGPLNSKNSSAKNNYGYCSYKASKRFSSLHAVLYFSPDFMSLGFMVWASGSMVFFLYRHKQQVQHNHSNRLSCRPSQEARATHTIMVLVSSFFVFYSVHSFLTIWTTVVANPGQWIVTNSVLVASCFPARSPFVLIMSDTHISQFCFACRTRKTLFPNLVVMP | |
| 63333141011011300000012263331463444144630302200110002301331332222003110000025333111000010013102210301231100000023211202200000000000102000000000020000000332202402230010001300010001020113000102033333233333210000012343210000001023112110001001000000011123102201134223333112100100000000002102200000000110334200000001020110010000000121420120020024352411300127 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHHHHHHHHHHHHHHHSSCCCCCHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCSSSSSSSSSHHHCCCHHHHHHHHHHHHSSSSCCCCHHHHHHHHHCCCSSSHHHHHHHHHHHHHHCCSSSSSSCCCCCCCCCCCCCCSSSSSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHCHHHHHCCCCCHHHHHHHHHCCCCCCCCSSSCC MVGDTLKLLSPLMTRYFFLLFYSTDSSDLNENQHPLDFDEMAFGKVKSGISFLIQTGVGILGNSFLLCFYNLILFTGHKLRPTDLILSQLALANSMVLFFKGIPQTMAAFGLKYLLNDTGCKFVFYYHRVGTRVSLSTICLLNGFQAIKLNPSICRWMEIKIRSPRFIDFCCLLCWAPHVLMNASVLLLVNGPLNSKNSSAKNNYGYCSYKASKRFSSLHAVLYFSPDFMSLGFMVWASGSMVFFLYRHKQQVQHNHSNRLSCRPSQEARATHTIMVLVSSFFVFYSVHSFLTIWTTVVANPGQWIVTNSVLVASCFPARSPFVLIMSDTHISQFCFACRTRKTLFPNLVVMP | |||||||||||||||||||||||||
| 1 | 4zwjA | 0.14 | 0.15 | 0.94 | 1.13 | Download | RTGTWDAYMCGTEGPNFYVPFSNATGVVRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQ-----KKLRPLNYILLNLAVADLFMVLGGFTSTLYTSLHGYFVFGPTGCNLQGFFATLGGEIALWSLVVLAIERYVVVCP---SNFRFGEN---HAIMGVAFTWVMALACAAPPLRYIPEGLQC-----SCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKEAAAQQQ-----ESATTQKAEKEVTRMVIIYVIAFLICWVPYASVAFYIFTHQGSGPIFMTIPAFFAKSAAIYNPVIYIMMNKQFRNCMLTTICCGKNVIFKKVSR | |||||||||||||||||||
| 2 | 4grvA | 0.14 | 0.15 | 0.79 | 1.98 | Download | --------------------------------NSDLDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLF----TLARKSLQSTVHYHLGSLALSDLLILLL-AMPVELYNFIWVHHFGDAGCRGYYFLRDACTYATALNVASLSVARYLAIC---HPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNRSADGTHPGGLVCTPIVDTATVKVVIQVNTFMSFLFPMLVISILNTVIANKLT------------VMSGSVQALRHGVLVARAVVIAFVVCWLPYHVRRLMFCYISDEQWYFYMLTNALAYASSAINPILLVSANFR---------------------- | |||||||||||||||||||
| 3 | 4zwjA | 0.11 | 0.15 | 0.93 | 1.35 | Download | QTPNRAKRVITTFRTGWDAYMCGTEGPNFSNATGVVRSYPQYYQFSMLAAYMFLLIVLGFPINFLTLYVTVQH---KKLRTPLNYILLNLAVADLFMVLGGFTSTLYTSLHGYFVFGPTGCNLQGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRF----GENHAIMGVAFTWVMALACAAPPSRYIPEG----------LQCSCGILKPENNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKEAAAQQ------QESATTQAEKEVTRMVIIYVIAFLICWVPYASVAFYIFTHQGSGPIFMTIPAFFAKSAAIYNPVIYIMMNKQFRNCMLTTICCGKNVIFKKVSR | |||||||||||||||||||
| 4 | 4djh | 0.13 | 0.23 | 0.81 | 1.52 | Download | ----------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVII---RYTKMKTATNIYIFNLALADALVTT--TMPFQSTVYMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVYIAVC-HPVKALD----FRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE-----DVDVIECSLQFPDDDDLFMKIVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFTDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS-AALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------- | |||||||||||||||||||
| 5 | 3odu | 0.11 | 0.20 | 0.85 | 1.27 | Download | ---------------------------------PCFREENANFNKIFLPTIYSIIFLTGIVGNGLVILVM----GYQKKRSMTDKYRLHLSVADLLFVIT--LPFWAVDAVANWYFGNFLCKAVHVIYTVNLYSSVWILAFISLDRYLAIVATNSQ-RPR---KLLAEKVVYVGVWIPALLLTIPDFIFANVSE-------ADDRYICDRFYPNDWVVVFQQHIMVGLILPGIVILSCYCIIISKLSHSGSNIFEMLRIAYGSKGHQKRKALKTTVILILAFFACWLPYYIGISIDSFILLEVHKWISITEALAFFHCCLNPILYAFLGAKFKTSAQHALTSGRPLEVLFQ-- | |||||||||||||||||||
| 6 | 4grvA | 0.13 | 0.15 | 0.80 | 1.42 | Download | --------------------------------NSDLDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLAR----KSLQSTVHYHLGSLALSDLLILLLAMPVELYNFIWHPWAFGDAGCRGYYFLRDACTYATALNVASLSVARYLAIC---HPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNRSADGTHPGGLVCTPIVDTATVKVVIQVNTFMSFLFPMLVISILNTVIANKLTVMSG------------SVQALRHGVLVARAVVIAFVVCWLPYHVRRLMFCYISDEQHYFYMLTNALAYASSAINPILYNLVSANFRQV------------------ | |||||||||||||||||||
| 7 | 4djh | 0.12 | 0.23 | 0.82 | 1.70 | Download | ------------------------------------------AIPVIITAVYSVVFVVGLVGNSLVMFV---IIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTP---LKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNITWDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS-AALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF------------ | |||||||||||||||||||
| 8 | 4buoA | 0.15 | 0.19 | 0.80 | 1.94 | Download | --------------------------------NSDLDVNTDIYSKVLVTAIYLALFVVGTVGNSVTL----FTLARKKSLSTVDYYLGSLALSDLLILLL-AMPVELYNFIWVWAFGDAGCRGYYFLRDACTYATALNVVSLSVELYLAICH------PFKAKTLMSTKKFISAIWLASALLAIPTMGLQNLSGDGTHP----GGLVCTPIVDATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQ---------PGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDEQWYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL------------- | |||||||||||||||||||
| 9 | 4grvA | 0.14 | 0.15 | 0.80 | 1.23 | Download | --------------------------------NSDLDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLA-----RKSLQTVHYHLGSLALSDLLILLLAMPVELYNFIWVHHAFGDAGCRGYYFLRDACTYATALNVASLSVARYLAIHPFKAK-----LMSRSRTKKFISAIWLASALLAIPMLFTMGLQNRSADGTHPGGLVCTPIVDTATVKVVIQVNTFMSFLFPMLVISILNTVIANKLTVM------------SGSVQALRHGVLVARAVVIAFVVCWLPYHVRRLMFCYISDEQWYFYMLTNALAYASSAINPILYNLVSANFRQV------------------ | |||||||||||||||||||
| 10 | 1l9hA | 0.15 | 0.19 | 0.92 | 1.17 | Download | MNG--------TEGPNFYVPFSNKTGVVRSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQ----HKKLRPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCPSNFRF------GENHAIMGVAFTWVMALACAAP-PLVGWSRYIPEGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLV----------FTVKEAAAATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTCGKNPLGDSTTVSK | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
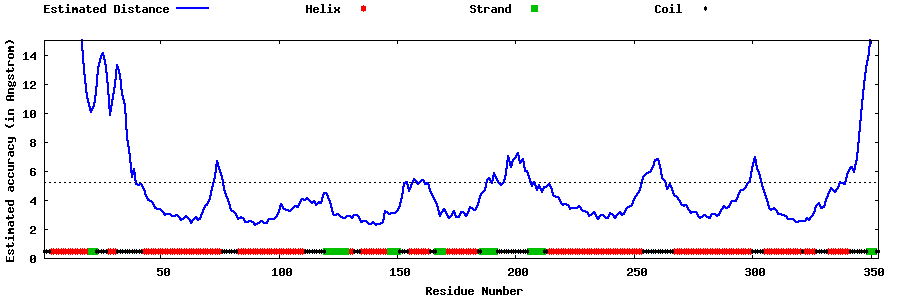 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
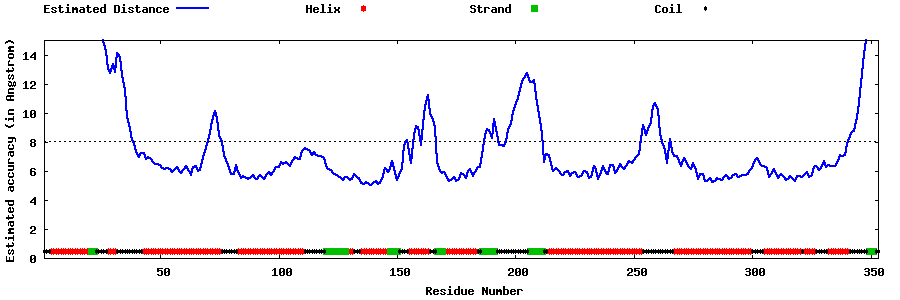 | |||
|
| |||
 | |||
|
| |||