

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | |
| | | | | | | | | | | | | | | | |
| MIPIQLTVFFMIIYVLESLTIIVQSSLIVAVLGREWLQVRRLMPVDMILISLGISRFCLQWASMLNNFCSYFNLNYVLCNLTITWEFFNILTFWLNSLLTVFYCIKVSSFTHHIFLWLRWRILRLFPWILLGSLMITCVTIIPSAIGNYIQIQLLTMEHLPRNSTVTDKLENFHQYQFQAHTVALVIPFILFLASTIFLMASLTKQIQHHSTGHCNPSMKARFTALRSLAVLFIVFTSYFLTILITIIGTLFDKRCWLWVWEAFVYAFILMHSTSLMLSSPTLKRILKGKC | |
| CCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCSSSSSSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHC | |
| 970489999999999999999998289986214998827978788899999999999999999999999996647999999999999949999999999999870110578769999998525119999999999999999999976630001210366885688986651379999999999999999999999999999999999725899999881999999999999999999999999999999854550999999999998277732877307758999987109 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | |
| | | | | | | | | | | | | | | | |
| MIPIQLTVFFMIIYVLESLTIIVQSSLIVAVLGREWLQVRRLMPVDMILISLGISRFCLQWASMLNNFCSYFNLNYVLCNLTITWEFFNILTFWLNSLLTVFYCIKVSSFTHHIFLWLRWRILRLFPWILLGSLMITCVTIIPSAIGNYIQIQLLTMEHLPRNSTVTDKLENFHQYQFQAHTVALVIPFILFLASTIFLMASLTKQIQHHSTGHCNPSMKARFTALRSLAVLFIVFTSYFLTILITIIGTLFDKRCWLWVWEAFVYAFILMHSTSLMLSSPTLKRILKGKC | |
| 723330332113113312331331022002000111045340320120000002110100111321100010133210100100012323300100000010000000323110000013304310011001133323010001113332313233354332111132334322112133333333333331331230002102320343343341131400220010012023333322301321011303452101010112002102300000011143014103748 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCSSSSSSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHC MIPIQLTVFFMIIYVLESLTIIVQSSLIVAVLGREWLQVRRLMPVDMILISLGISRFCLQWASMLNNFCSYFNLNYVLCNLTITWEFFNILTFWLNSLLTVFYCIKVSSFTHHIFLWLRWRILRLFPWILLGSLMITCVTIIPSAIGNYIQIQLLTMEHLPRNSTVTDKLENFHQYQFQAHTVALVIPFILFLASTIFLMASLTKQIQHHSTGHCNPSMKARFTALRSLAVLFIVFTSYFLTILITIIGTLFDKRCWLWVWEAFVYAFILMHSTSLMLSSPTLKRILKGKC | |||||||||||||||||||||||||
| 1 | 4djhA | 0.12 | 0.17 | 0.94 | 1.44 | Download | --SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR----TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKA----LDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDYSWWDLFMKICVFIFAVIPVLIIIVCYTLMILRLKSVRLLSD--------RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFC | |||||||||||||||||||
| 2 | 5tjvA | 0.09 | 0.20 | 0.92 | 2.56 | Download | VLNPSQQLAIAVLSLTLGTFTVLENLLVL----CVILHSRSLRPSYHFIGSLAVADLLGSVIFVYSFIDFHVDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLA----YKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHID---------ETYLMFWIGVTSVLLLFIVYAYMYILWKAGKRAMSFSD-----QARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSM- | |||||||||||||||||||
| 3 | 4xt1A | 0.10 | 0.17 | 0.91 | 2.11 | Download | DVLNQSKPVTLFLYGVVFLFGSIGNFLVIFTITW---RRRIQCSGDVYFINLAAADLLFVCT--LPLWMQYLLDSVPCTLLTACFYVAMFASLCFITEIALDRYYAIVYMRYRP--------VKQACLFSIFWWIFAVIIAIPHFMVVTKKDNQCMTDYDYL------EVSYPIILNVELMLGAFVIPLSVISYCYYRISRIVA-------VSQSRHKGRIVRVLIAVVLVFIIFWLPYHLTLFVDTLKLERSLKRALILTESLAFCHCCLNPLLYVFVGTKFRQELHCLL | |||||||||||||||||||
| 4 | 4djh | 0.12 | 0.22 | 0.97 | 1.58 | Download | --SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR---YTKMKTATNIYIFNLALADALVTTTMPFQSTVYLSWPDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVC---HPVKAL-DFRTPLKAKIINICIWLLSSSVGISAIVLGGTKREDVDVIECSLQFPDDDY-SWWDLFMKIVFIFAFVIPVLIIIVCYTLMILRLSVRLLSGNIDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEAGAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFC | |||||||||||||||||||
| 5 | 5glh | 0.12 | 0.19 | 0.92 | 1.19 | Download | EIKETFKYINTVVSCLVFVLGIIGNSTLLYIIYK--------NGPNILIASLALGDLLHIVIAIPINVYKLLAGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASW--------------WTAVEIVLIWVVSVVLAVPEAIGFDIITMDY-KGSYLRICLLHPVQKTAQFYDWWLFSFYFCLPLAITAFFYTLMTCEMLRKNEGLRDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARIKLTLYNNVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSAL | |||||||||||||||||||
| 6 | 4djhA | 0.12 | 0.17 | 0.92 | 1.52 | Download | ---PAIPVIITAVYSVVFVVGLVGNSLVMFVIIRY---TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKA----LDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR-----EDVDVIECSLQFWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSD--------RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFC | |||||||||||||||||||
| 7 | 3uon | 0.11 | 0.23 | 0.47 | 1.70 | Download | ------VVFIVLVAGSLSLVTIIGNILVMVSIKV---NRHLQTVNNYFLFSLACADLIIGVFSMNLYTGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQ-------------------------------------------------------------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4buoA | 0.14 | 0.17 | 0.96 | 2.27 | Download | NTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLAR----KKSLSTVDYYLGSLALSDLLILLLAMPVELYNFIWGDAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIP-----MLFTMGLQNLSGDGTHPGGLVCTPIVDTLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQP--GRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYILFDFYHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL | |||||||||||||||||||
| 9 | 4djhA | 0.13 | 0.17 | 0.92 | 1.39 | Download | --SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR----TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVC---HPVKA-LDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDV-----IECSLQFWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSD--------RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSASSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFC | |||||||||||||||||||
| 10 | 4ea3A | 0.11 | 0.20 | 0.92 | 1.44 | Download | --PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILR---HTKMKTATNIYIFNLALADTLVLLTLPFQGTDILLGFNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAIC---HPT--------SSKAQAVNVAIWALASVVGVPVAIMGSAQVEDEEIECLVEI--PTPQDYWGPVFAICIFLFSFIVPVLVISVCYSLMIRRLRGVRLLSGSREKD---RNLRRITRLVLVVVAVFVGCWTPVQVFVLAQGLGVQAILRFCTALGYVNSCLNPILYAFLDENFKACFR--- | |||||||||||||||||||
| ||||||||||||||||||||||||||
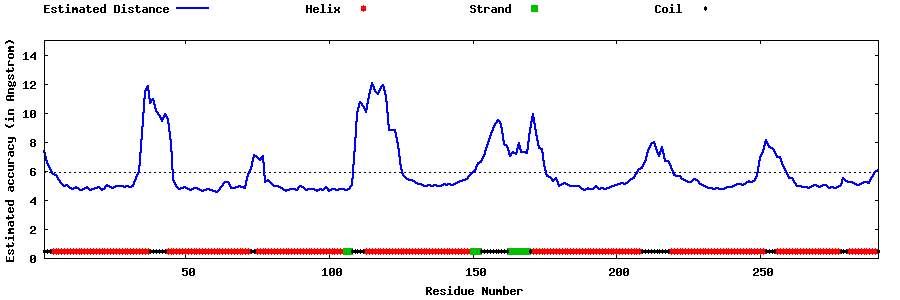
| Generated 3D models | Estimated local accuracy of models | ||
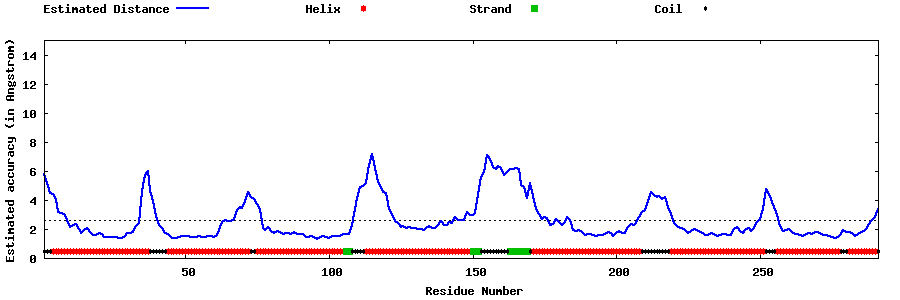 | |||
|
|
|||
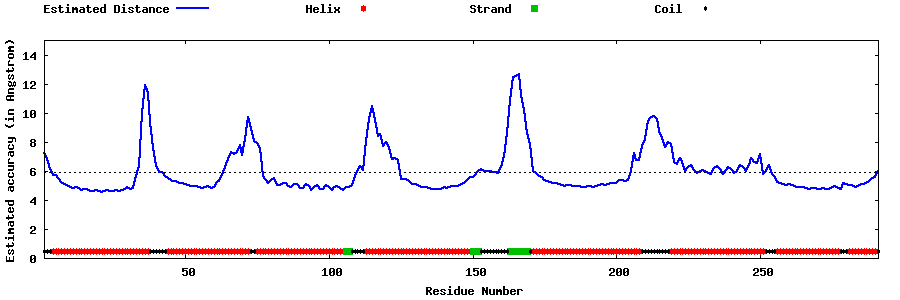 | |||
|
| |||
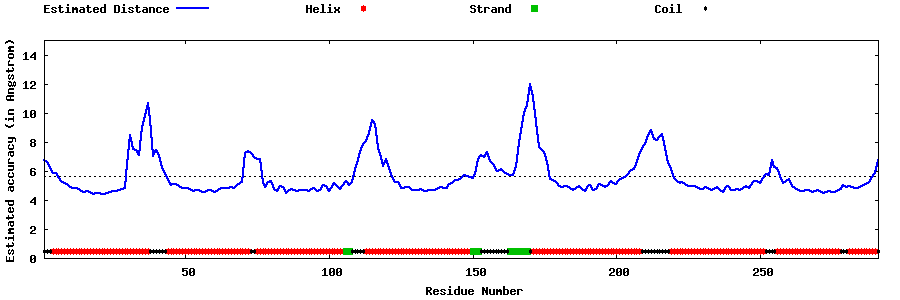 | |||
|
| |||
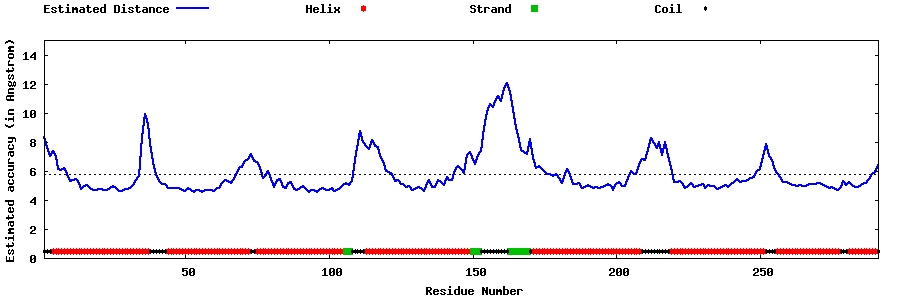 | |||
|
| |||
 | |||
|
| |||