

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MFSPADNIFIILITGEFILGILGNGYIALVNWIDWIKKKKISTVDYILTNLVIARICLISVMVVNGIVIVLNPDVYTKNKQQIVIFTFWTFANYLNMWITTCLNVFYFLKIASSSHPLFLWLKWKIDMVVHWILLGCFAISLLVSLIAAIVLSCDYRFHAIAKHKRNITEMFHVSKIPYFEPLTLFNLFAIVPFIVSLISFFLLVRSLWRHTKQIKLYATGSRDPSTEVHVRAIKTMTSFIFFFFLYYISSILMTFSYLMTKYKLAVEFGEIAAILYPLGHSLILIVLNNKLRQTFVRMLTCRKIACMI | |
| CCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCSSSSSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHCCSSSC | |
| 997899999999999999999999999999999999178788788999999999899999999716725743413025478999999999983999999999999971786089976999999986615999999999999999999997515432442000589833677654407899999999999999999999999999999999999974889999999808999999999999999999998999999999707768999999999999688877999508759999999999872374439 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MFSPADNIFIILITGEFILGILGNGYIALVNWIDWIKKKKISTVDYILTNLVIARICLISVMVVNGIVIVLNPDVYTKNKQQIVIFTFWTFANYLNMWITTCLNVFYFLKIASSSHPLFLWLKWKIDMVVHWILLGCFAISLLVSLIAAIVLSCDYRFHAIAKHKRNITEMFHVSKIPYFEPLTLFNLFAIVPFIVSLISFFLLVRSLWRHTKQIKLYATGSRDPSTEVHVRAIKTMTSFIFFFFLYYISSILMTFSYLMTKYKLAVEFGEIAAILYPLGHSLILIVLNNKLRQTFVRMLTCRKIACMI | |
| 734333321122133123313310220021011100443403100100000021200000013331000000031134330000010111231130010000000000000012311000101220431001203312332232332130303232333334434210112133432210111113333333333213323320210022013303433443411314102200200120221033113111100012133442001010133002000100101023143013001200212303016 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCSSSSSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHCCSSSC MFSPADNIFIILITGEFILGILGNGYIALVNWIDWIKKKKISTVDYILTNLVIARICLISVMVVNGIVIVLNPDVYTKNKQQIVIFTFWTFANYLNMWITTCLNVFYFLKIASSSHPLFLWLKWKIDMVVHWILLGCFAISLLVSLIAAIVLSCDYRFHAIAKHKRNITEMFHVSKIPYFEPLTLFNLFAIVPFIVSLISFFLLVRSLWRHTKQIKLYATGSRDPSTEVHVRAIKTMTSFIFFFFLYYISSILMTFSYLMTKYKLAVEFGEIAAILYPLGHSLILIVLNNKLRQTFVRMLTCRKIACMI | |||||||||||||||||||||||||
| 1 | 4djhA | 0.11 | 0.20 | 0.91 | 1.40 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR----TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKA----LDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSDR-----------NLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------- | |||||||||||||||||||
| 2 | 5tjvA | 0.10 | 0.17 | 0.89 | 2.56 | Download | LNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHS--RSLRCRPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLA----YKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHID----------ETYLMFWIGVTSVLLLFIVYAYMYILWKAGKRAMSFSD--------QARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------- | |||||||||||||||||||
| 3 | 4n6hA | 0.10 | 0.22 | 0.92 | 2.23 | Download | SSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY--TKMK-TATNIYIFNLALADALATST--LPFQSAKYLMETWFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKA----LDFRTPAKAKLINICIWVLASGVGVPIMVMA-------VTRPRDGAVVCMLQFPYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV-----RLLSGSK-EKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---- | |||||||||||||||||||
| 4 | 4ib4 | 0.14 | 0.21 | 0.93 | 1.52 | Download | EQGNKLHWAALLILMVIIPTIGGNTLVILAVSLE---KKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIK---KPIQAN-QYNSRATAFIKITVVWLISIGIAIPVPIKGITNPNN--I-TCVLTKE---R--FGDFML--FGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR---- | |||||||||||||||||||
| 5 | 5glh | 0.14 | 0.19 | 0.89 | 1.18 | Download | IKETFKYINTVVSCLVFVLGIIGNSTLLYIIYK--------NGPNILIASLALGDLLHIVIAIPINVYKLLAEDWPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVAS---W-----------WTAVEIVLIWVVSVVLAVPEAIGFDIITMDY--KGSYLRICLLHPVQKFMQFYDWWLFSFYFCLPLAITAFFYTLMTCEMLRKNEGLRLTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKTLYNNLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSAL--------- | |||||||||||||||||||
| 6 | 4grvA | 0.11 | 0.20 | 0.88 | 1.50 | Download | --IYSKVLVTAIYLALFVVGTVGNSVTLFTLA----RKSLQSTVHYHLGSLALSDLLILLLAMPVELYNFIWHPWAFGDAGCRGYYFLRDACTYATALNVASLSVARYLAICH----PFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNRSADGTHPGGLVCTPIVDTATVKVVIQVNTFMSFLFPMLVISILNTVIANKLTVM------------SGSVQALRHGVLVARAVVIAFVVCWLPYHVRRLMFCYISYHYFYMLTNALAYASSAINPILYNLVSANFRQV-------------- | |||||||||||||||||||
| 7 | 3uon | 0.09 | 0.19 | 0.92 | 1.69 | Download | -----VVFIVLVAGSLSLVTIIGNILVMVSIKV---NRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIA----AAWVLSFILWAPAILFWQFIVGVRTVEDGECY--IQFFS-N--AAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFAADAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAP-CIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM-------- | |||||||||||||||||||
| 8 | 4n6hA | 0.09 | 0.22 | 0.91 | 2.45 | Download | SSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY---TKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVK----ALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTPRD---------GAVVCMLQFYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV------RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKCG----- | |||||||||||||||||||
| 9 | 4djhA | 0.12 | 0.20 | 0.89 | 1.38 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR----TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVC---HPVKA-LDFRTPLKAKIINICIWLLSSSVGISAIVL----GGTKVREDVDVIECSLQFPDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLL------------DRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------- | |||||||||||||||||||
| 10 | 3aymA | 0.13 | 0.20 | 0.95 | 1.63 | Download | VPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTK----TKSLTPANMFIINLAFSDFTFSLVNGFPLMTISFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGPMAASKKMS-----HRRAFIMIIFVWLWSVLWAIGPI-----FGWGAYTLEGVLCNCSFDYISRTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNAKEANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQF | |||||||||||||||||||
| ||||||||||||||||||||||||||
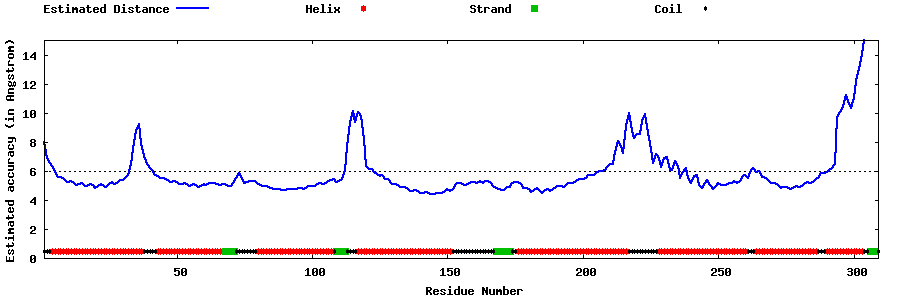
| Generated 3D models | Estimated local accuracy of models | ||
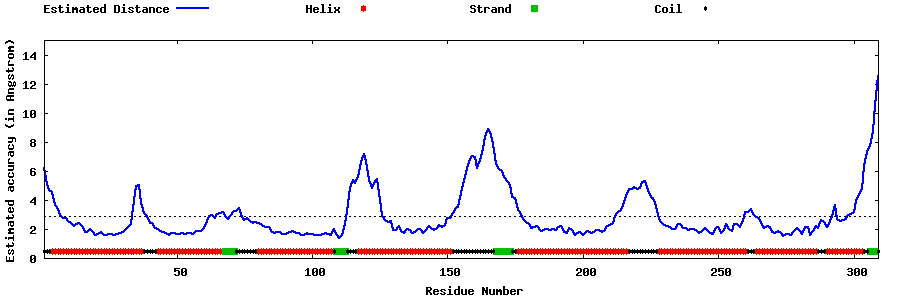 | |||
|
|
|||
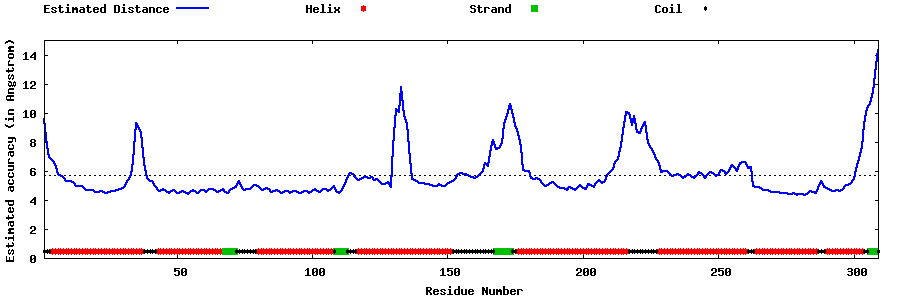 | |||
|
| |||
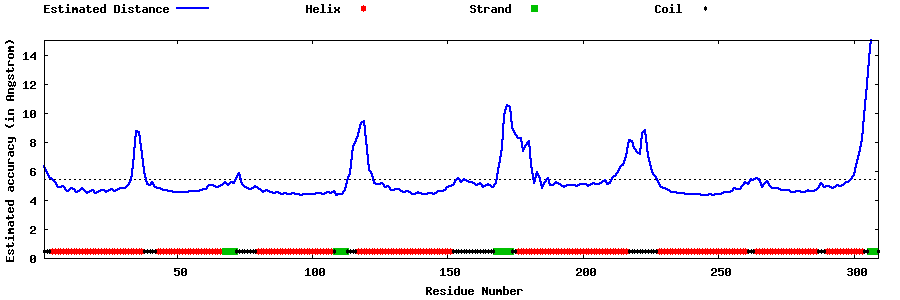 | |||
|
| |||
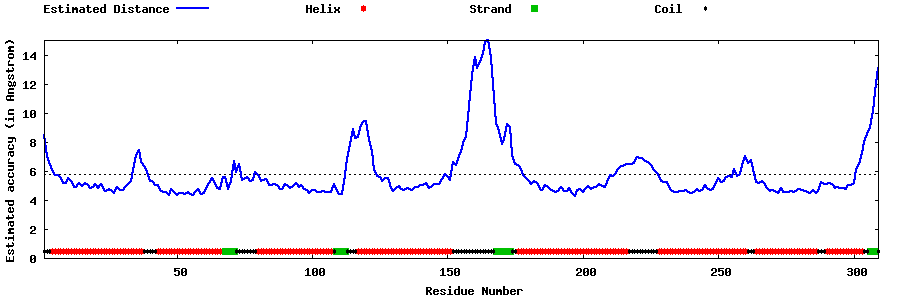 | |||
|
| |||
 | |||
|
| |||