

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MFVASERKMRAHQVLTFLLLFVITSVASENASTSRGCGLDLLPQYVSLCDLDAIWGIVVEAVAGAGALITLLLMLILLVRLPFIKEKEKKSPVGLHFLFLLGTLGLFGLTFAFIIQEDETICSVRRFLWGVLFALCFSCLLSQAWRVRRLVRHGTGPAGWQLVGLALCLMLVQVIIAVEWLVLTVLRDTRPACAYEPMDFVMALIYDMVLLVVTLGLALFTLCGKFKRWKLNGAFLLITAFLSVLIWVAWMTMYLFGNVKLQQGDAWNDPTLAITLAASGWVFVIFHAIPEIHCTLLPALQENTPNYFDTSQPRMRETAFEEDVQLPRAYMENKAFSMDEHNAALRTAGFPNGSLGKRPSGSLGKRPSAPFRSNVYQPTEMAVVLNGGTIPTAPPSHTGRHLW | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHCCCCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHSSSSSSSSCCCCCCSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCSSSSSSSCHHHCCCHHHHCCCCCCCCCCCCCCCCCCCCCSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSSSSCCCCCCCCCCCHHCCCCC | |
| 9769888667999977108899987878898889999988896414531624159999999999999999999999987600012245445257899999999999999999854765144628899999999999999999887577440657695122005547777753531050278962675431204679678979999999999999999998743667323410999999999999999999988620133200103688899999999999999998750158999835242471444047888665643467765675400488878677643557887766676667764467888876778888986168851698779999751022469 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MFVASERKMRAHQVLTFLLLFVITSVASENASTSRGCGLDLLPQYVSLCDLDAIWGIVVEAVAGAGALITLLLMLILLVRLPFIKEKEKKSPVGLHFLFLLGTLGLFGLTFAFIIQEDETICSVRRFLWGVLFALCFSCLLSQAWRVRRLVRHGTGPAGWQLVGLALCLMLVQVIIAVEWLVLTVLRDTRPACAYEPMDFVMALIYDMVLLVVTLGLALFTLCGKFKRWKLNGAFLLITAFLSVLIWVAWMTMYLFGNVKLQQGDAWNDPTLAITLAASGWVFVIFHAIPEIHCTLLPALQENTPNYFDTSQPRMRETAFEEDVQLPRAYMENKAFSMDEHNAALRTAGFPNGSLGKRPSGSLGKRPSAPFRSNVYQPTEMAVVLNGGTIPTAPPSHTGRHLW | |
| 7230353304446313000300415334434630450345212410300226100000010301311220220000000201102035442100000000211331110000000112300000002110301000000000000000000113344332100000000032022000000000123413130323210000001110000220221002000341462220000000001202001200100001043323312403200000000010111010111100000002154433653245443433443354446343332424223374344435533333433644446544544444345433331101112445323643223003327 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHCCCCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHSSSSSSSSCCCCCCSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCSSSSSSSCHHHCCCHHHHCCCCCCCCCCCCCCCCCCCCCSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSSSSCCCCCCCCCCCHHCCCCC MFVASERKMRAHQVLTFLLLFVITSVASENASTSRGCGLDLLPQYVSLCDLDAIWGIVVEAVAGAGALITLLLMLILLVRLPFIKEKEKKSPVGLHFLFLLGTLGLFGLTFAFIIQEDETICSVRRFLWGVLFALCFSCLLSQAWRVRRLVRHGTGPAGWQLVGLALCLMLVQVIIAVEWLVLTVLRDTRPACAYEPMDFVMALIYDMVLLVVTLGLALFTLCGKFKRWKLNGAFLLITAFLSVLIWVAWMTMYLFGNVKLQQGDAWNDPTLAITLAASGWVFVIFHAIPEIHCTLLPALQENTPNYFDTSQPRMRETAFEEDVQLPRAYMENKAFSMDEHNAALRTAGFPNGSLGKRPSGSLGKRPSAPFRSNVYQPTEMAVVLNGGTIPTAPPSHTGRHLW | |||||||||||||||||||||||||
| 1 | 4or2A | 0.22 | 0.15 | 0.60 | 1.68 | Download | ---------------------------------------------VRYLEWSNIESIIAIAFSCLGILVTLFVTLIFVLYTPVVKSSS----RELCYIILAGIFLGYVCPFTLIAKPTTTSCYLQRLLVGLSSAMCYSALVTKTNRIARILRKPRFMSAWAQVIIASILISVQLTLVVTLIIMEPPMEVYLICNTSNLGVVAPLGYNGLLIMSCTYYAFKTR--NVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSN--------YKIITTCFAVSLSVTVALGCMFTPKMYIIIAKPE------------------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 2 | 5l7dA | 0.08 | 0.18 | 0.93 | 1.74 | Download | FLRCTPDRFPEGCTNEVQNIKFNSSGQCEVPLVRTDNPKSWYEDVEGCGLFTEAEHQDMHSYIAAFGAVTGLCTLFTLATFVADWRNSNRYPAVILFYVNACFFVGSIGWLAQFMDGARREIVVIVYYALMAGFVWFVVLTYAWHTSFKALQPLSGKTSYFHLLTWSLPFVLTVAILAVAQVDGDSVSGICFVGYKNYRYRAGFVLAPIGLVLIVGGYFLIRGVMTLFSARRQLADLEDNWETLNDNLKMRAAALDAQKATPPSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLERARSTLSKINEFFNQAEWERSFRDYVLCQANVTIGLPTKQP----------------------------IPDCEIKNRPSLLVCRLT | |||||||||||||||||||
| 3 | 4or2A | 0.21 | 0.15 | 0.60 | 2.95 | Download | ---------------------------------------------VRYLEWSNIESIIAIAFSCLGILVTLFVTLIFVLDTPVVKSSSRE----LCYIILAGIFLGYVCPFTLIAKPTTTSCYLQRLLVGLSSAMCYSALVTKTNRIARILAKPRFMSAWAQVIIASILISVQLTLVVTLIIMEPPKEVYLICNTSNLGVVAPLGYNGLLIMSCTYYAFKT--RNVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSNY--------KIITTCFAVSLSVTVALGCMFTPKMYIIIAKPERN----------------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 4 | 6eznF | 0.08 | 0.22 | 1.00 | 1.74 | Download | LFAVIKFESIIHEFNWFDDRTWYPLGRVTGGTLYPGRNWLGLPIDIRNVCVLFAPLFSGVTAWATYEFTITLLMVTFMFWIKAQKTGSIMHATCAAYVFITNLIPLHVFLLILMGRYSSKLYSAYTTWYAIGTAALGVFGLIQIVAFGDFVKGQISTAKFKVIMQPVSWPAFFFDTHFLIWLFPAGVFLLFLDLKDEHVFVIAYSTPVICVSAAVALSKIFDIYLDFKIKPAALLAKLIVSGSFIFYLYLFVFHSLVDNNTWNNTHIAIVGKAMASPEEKSYEILKEHDVDYVLVIFGGLIGFGRISEGIWPEEIKERDFYTAEGEYRVDARASETYKDFPQLFNGGQATDRVRQQMITPLDVPPLDYKKDDAQGRTLRDVGELTRSSTKTRRSIKRPELGLV | |||||||||||||||||||
| 5 | 4or2A | 0.22 | 0.15 | 0.60 | 3.48 | Download | ---------------------------------------------VRYLEWSNIESIIAIAFSCLGILVTLFVTLIFVLYTPVVKSSSR----ELCYIILAGIFLGYVCPFTLIAKPTTTSCYLQRLLVGLSSAMCYSALVTKTNRIARILRKPRFMSAWAQVIIASILISVQLTLVVTLIIMEPPMEVYLICNTSNLGVVAPLGYNGLLIMSCTYYAFKT--RNVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSNYK--------IITTCFAVSLSVTVALGCMFTPKMYIIIAKPE------------------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 6 | 3orgA | 0.08 | 0.19 | 0.89 | 1.29 | Download | ICAIGGGLPVGWEGPNVHIACIIAHQFYRLGVFKELCTD------------RALRLQTLAAACAVGLASSFGVLYSIETIASFYLVQ------------AFWKGVLSALSGAIVYELDVRTQTLLYAILGALMGVLGALFIRCVRSIYELRMRHYPGTNRYFLVGVVALFASALQYP------FPRATINDLFKAVTIKFILVALSISFLIGAGFGRLYGELMRVVFGNAIVGSYAVVGAAAFTAGVTSCAVIIFEVTGQIRHLVPVLISVLLAVIVGNAFNTLVLMKHLPYMPILRR----------DRSPEMTAREIMHPIEGEPHLFPDSEPQHIKGILEKFPNRLPVIDANGYLLGKEIVDRLQHVVVPCDVSPI---VVTSYSLVRQLHFLFVMLMPS | |||||||||||||||||||
| 7 | 4or2A | 0.22 | 0.15 | 0.60 | 5.93 | Download | ---------------------------------------------VRYLEWSNIESIIAIAFSCLGILVTLFVTLIFVLDTPVVKS----SSRELCYIILAGIFLGYVCPFTLIAKPTTTSCYLQRLLVGLSSAMCYSALVTKTNRIARILARKPFMSAWAQVIIASILISVQLTLVVTLIIMEPPMPVYLICNTSNLGVVAPLGYNGLLIMSCTYYAFKT--RNVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSNY--------KIITTCFAVSLSVTVALGCMFTPKMYIIIA-KPE------------------------------------------------------------------------------------------------------ | |||||||||||||||||||
| 8 | 4oo9 | 0.19 | 0.21 | 0.61 | 4.52 | Download | -------------------------------------------SPVQYLRWGDPAPIAAVVFACLGLLATLFVTVVFYRDTPVVKSSSRE----LCYIILAGICLGYLCTF-LIAKPKQIYCYLQRIGIGLSPAMSYSALVTKTYRAARILAMSKKNSSAAQLVIAFILICIQLGIIVALFIMEPPDIVYLICNTTNLGVVAPLGYNGLLILACTFYAFKT--RNVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSNYKII--------TMCFSVSLSATVALGCMFVPKVYIILAKPERN----------------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 9 | 4or2A | 0.22 | 0.15 | 0.60 | 2.41 | Download | ---------------------------------------------VRYLEWSNIESIIAIAFSCLGILVTLFVTLIFVLYTPVVKSSS----RELCYIILAGIFLGYVCPFTLIAKPTTTSCYLQRLLVGLSSAMCYSALVTKTNRIARILRKPRFMSAWAQVIIASILISVQLTLVVTLIIMEPPKEVYLICNTSNLGVVAPLGYNGLLIMSCTYYAFKT--RNVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSN--------YKIITTCFAVSLSVTVALGCMFTPKMYIIIAKPE------------------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 10 | 4or2 | 0.22 | 0.17 | 0.61 | 6.71 | Download | ---------------------------------------LIPVRYL---EWSNIESIIAIAFSCLGILVTLFVTLIFVL--YRDTPVVKSSSRELCYIILAGIFLGYVCPFTLIAKPTTTSCYLQRLLVGLSSAMCYSALVTKTNRIARILAKPRFMSAWAQVIIASILISVQLTLVVTLIIMEPPMPIYLICNTSNLGVVAPLGYNGLLIMSCTYYAFKT--RNVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSNYK--------IITTCFAVSLSVTVALGCMFTPKMYIIIAKPE------------------------------------------------------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
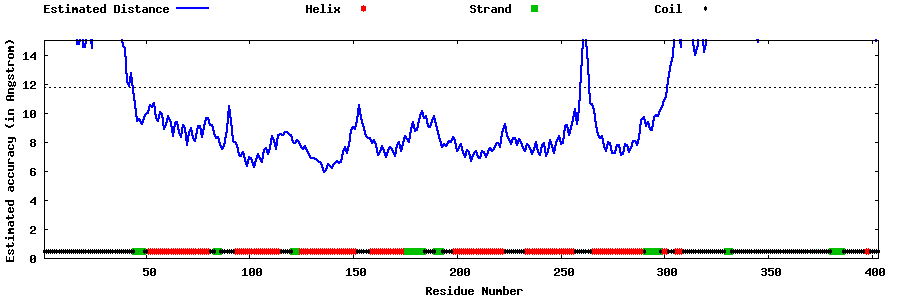
| Generated 3D models | Estimated local accuracy of models | ||
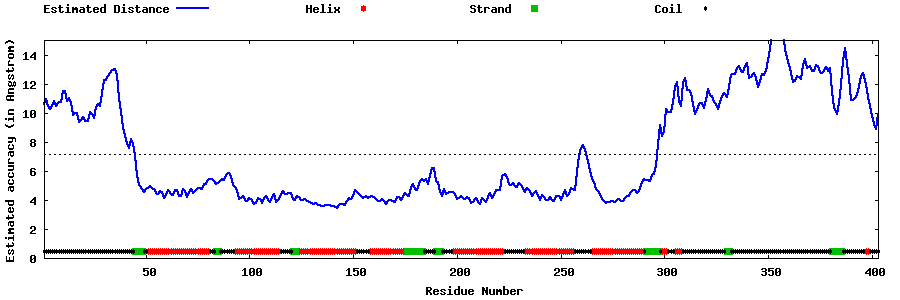 | |||
|
|
|||
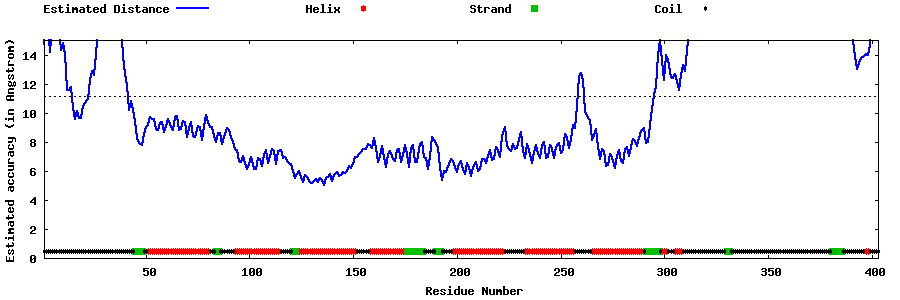 | |||
|
| |||
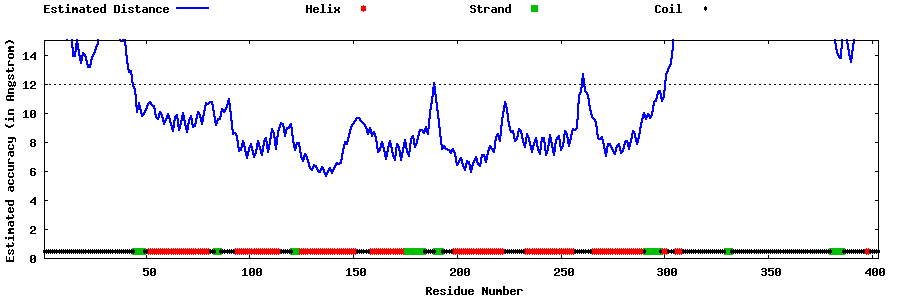 | |||
|
| |||
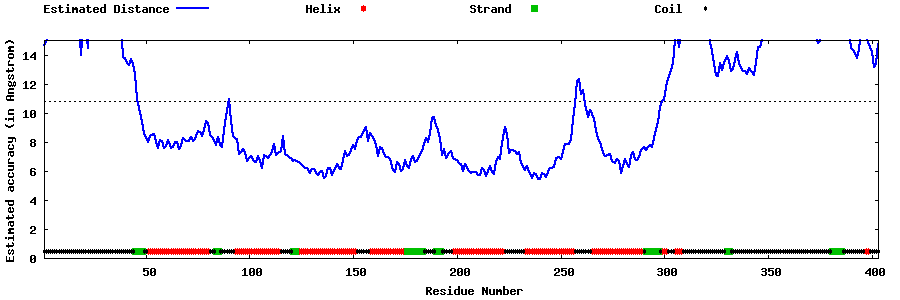 | |||
|
| |||
 | |||
|
| |||