

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 | |
| | | | | | | | | | | | | | | | | | | | | | | | |
| MERAPPDGPLNASGALAGEAAAAGGARGFSAAWTAVLAALMALLIVATVLGNALVMLAFVADSSLRTQNNFFLLNLAISDFLVGAFCIPLYVPYVLTGRWTFGRGLCKLWLVVDYLLCTSSAFNIVLISYDRFLSVTRAVSYRAQQGDTRRAVRKMLLVWVLAFLLYGPAILSWEYLSGGSSIPEGHCYAEFFYNWYFLITASTLEFFTPFLSVTFFNLSIYLNIQRRTRLRLDGAREAAGPEPPPEAQPSPPPPPGCWGCWQKGHGEAMPLHRYGVGEAAVGAEAGEATLGGGGGGGSVASPTSSSGSSSRGTERPRSLKRGSKPSASSASLEKRMKMVSQSFTQRFRLSRDRKVAKSLAVIVSIFGLCWAPYTLLMIIRAACHGHCVPDYWYETSFWLLWANSAVNPVLYPLCHHSFRRAFTKLLCPQKLKIQPHSSLEHCWK | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCC | |
| 9978999987677787888777788888888999999999999999999957862434683265575559999999999999999842499999972811386389999999999999999999999988503124462226100664889899999999999999978987201147876567888630144540799999999999989999999999999999998876411332233334444344566654445543356775432223465545666432334444333333335655565443222223320001110122201101344543332000012457777899999999999997308999999998726656865999999999999853437889875899999999996756578999861023469 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 | |
| | | | | | | | | | | | | | | | | | | | | | | | |
| MERAPPDGPLNASGALAGEAAAAGGARGFSAAWTAVLAALMALLIVATVLGNALVMLAFVADSSLRTQNNFFLLNLAISDFLVGAFCIPLYVPYVLTGRWTFGRGLCKLWLVVDYLLCTSSAFNIVLISYDRFLSVTRAVSYRAQQGDTRRAVRKMLLVWVLAFLLYGPAILSWEYLSGGSSIPEGHCYAEFFYNWYFLITASTLEFFTPFLSVTFFNLSIYLNIQRRTRLRLDGAREAAGPEPPPEAQPSPPPPPGCWGCWQKGHGEAMPLHRYGVGEAAVGAEAGEATLGGGGGGGSVASPTSSSGSSSRGTERPRSLKRGSKPSASSASLEKRMKMVSQSFTQRFRLSRDRKVAKSLAVIVSIFGLCWAPYTLLMIIRAACHGHCVPDYWYETSFWLLWANSAVNPVLYPLCHHSFRRAFTKLLCPQKLKIQPHSSLEHCWK | |
| 8443427232322232334433444343231000000002112002201311200000001133011100000000010000000000120000001230210310001000000000000010000000000100020030333324200000000000200111010000001114444414644120202332000000021013000000020011001000220333244344343433332223222334424433343332333233333443444333342413443332332222222222222122222222223233444334433433323222222244433303200000001020120000100010003531013100000013013001100000000053013002200202234243454344128 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 | | | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCC MERAPPDGPLNASGALAGEAAAAGGARGFSAAWTAVLAALMALLIVATVLGNALVMLAFVADSSLRTQNNFFLLNLAISDFLVGAFCIPLYVPYVLTGRWTFGRGLCKLWLVVDYLLCTSSAFNIVLISYDRFLSVTRAVSYRAQQGDTRRAVRKMLLVWVLAFLLYGPAILSWEYLSGGSSIPEGHCYAEFFYNWYFLITASTLEFFTPFLSVTFFNLSIYLNIQRRTRLRLDGAREAAGPEPPPEAQPSPPPPPGCWGCWQKGHGEAMPLHRYGVGEAAVGAEAGEATLGGGGGGGSVASPTSSSGSSSRGTERPRSLKRGSKPSASSASLEKRMKMVSQSFTQRFRLSRDRKVAKSLAVIVSIFGLCWAPYTLLMIIRAACHGHCVPDYWYETSFWLLWANSAVNPVLYPLCHHSFRRAFTKLLCPQKLKIQPHSSLEHCWK | |||||||||||||||||||||||||
| 1 | 5wiuA | 0.23 | 0.23 | 0.84 | 2.47 | Download | ---------------------------------GAAALVGGVLLIGAVLAGNSLVCVSVATERALQTPTNSFIVSLAAADLLLALLVLPLFVYSEVQGAWLLSPRLCDALMAMDVMLCTASIFNLCAISVDRFVAVAVPLRYNRQGGS-RRQLLLIGATWLLSAAVAAPVLCGLN----------DPAVCRL-EDRDYVVYSSVCSFFLPCPLMLLLYWATFRGLQRWEVARRADLEDNWETLNDNLK---------VIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLAKITGRERKAMRVLPVVVGAFLLCWTPFFVVHITQALCPACSVPPRLVSAVTWLGYVNSALNPVIYTVFNAEFRNVFRKA------------------- | |||||||||||||||||||
| 2 | 4iarA | 0.23 | 0.24 | 0.84 | 4.64 | Download | ----------------------------ISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRT-PKRAAVMIALVWVFSISISLPPFFWRQ----------ASECVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKAT--------------PPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLAARERKATKTLGIILGAFIVCWLPFFIISLVMPIW----FHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--------------- | |||||||||||||||||||
| 3 | 5uenA | 0.22 | 0.22 | 0.84 | 3.52 | Download | ----------------------------SISAFQAAYIGIEVLIALVSVPGNVLVIWAVKVNQALRDATFCFIVSLAVADVAVGALVIPLAILINIGPQTYF--HTCLMVACPVLILTQSSILALLAIAVDRYLRVKIPLRYKMVVTP-RRAAVAIAGCWILSFVVGLTPMFGWNNWAAAGSMGEPVIKCEVISMEYMVYFNFFVWVLPPLLLMVLIYLEVFYLIRKQLADLEDNWETLNDNV------------------------------------KDALTKMRAAALDAPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYQKYLERARSTLQKELKIAKSLALILFLFALSWLPLHILNCITLFCPSCHKPSILTYIAIFLTHGNSAMNPIVYAFRIQKFRVTFLKIWNDHFRCQPLEVLF----- | |||||||||||||||||||
| 4 | 3uon | 0.24 | 0.25 | 0.89 | 1.62 | Download | ------------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKR-TTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVEDGECYIQFFSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMLRIDEGLRLKIYKDTEGGHLLTKSPSLNAADKAIGRNTNGVITKDNRGILRNAK-LKPVYDSLDAVALIETGVAGNSLLQQKRWDVNLAKSRWYNQTNKRVITTFRTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCA-PCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM----------------- | |||||||||||||||||||
| 5 | 3uon | 0.24 | 0.25 | 0.89 | 1.25 | Download | ------------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRT-TKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVEDGECYIQFFSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMLRIDEGLRLKIYKDTEGIGHLLTKSPSLNAAKAIGRNTNGVITKEKLVRGILRNAKDSLDAVRRAALINQMGEAGSLRMLQQKRDEAAVNLAKSTPNRKRVITTFRTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAP-CIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM----------------- | |||||||||||||||||||
| 6 | 5wiuA | 0.24 | 0.23 | 0.84 | 3.32 | Download | ---------------------------------GAAALVGGVLLIGAVLAGNSLVCVSVATERALQTPTNSFIVSLAAADLLLALLVLPLFVYSEVQGAWLLSPRLCDALMAMDVMLCTASIFNLCAISVDRFVAVAVPLRYNRQGG-SRRQLLLIGATWLLSAAVAAPVLCGLN-----------DPAVCRLEDRDYVVYSSVCSFFLPCPLMLLLYWATFRGLQRWEVARRADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPPEMKDFRHGFDILVGQIDDALKLANEGK---------VKEAQAAAEQLKTTRNAYIQKYLAKITGRERKAMRVLPVVVGAFLLCWTPFFVVHITQALCPACSVPPRLVSAVTWLGYVNSALNPVIYTVFNAEFRNVFRKA------------------- | |||||||||||||||||||
| 7 | 3uon | 0.24 | 0.25 | 0.88 | 1.77 | Download | ---------------------------------VVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKR-TTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVEDGECYIQFFSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMLRIDEGLRLKIYKDTEGGGHLLTKSPSLNAAKAIGRNTNGVITKDAEKLFNDVRGILRNAKLKPVYDLDAVRAGMLQQKRWDEAAVNLAKSRWYNQTNRAKRTTFRTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAP-CIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM----------------- | |||||||||||||||||||
| 8 | 3uonA | 0.25 | 0.25 | 0.89 | 4.40 | Download | ---------------------------TFE---VVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRT-TKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVEDGECYIQFFSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMLRIDEAAKSELDKAIGRNTNGVITKDEAEKLFNQDVDAAVRGILRNAKLKPVYDSLDAVRRAALINMVFQMGETGVAGFTNSLRMLQQKRWDEAAVNLAKSRWYNQTPNRAKRVITTFRTGVTRTILAILLAFIITWAPYNVMVLINTFCA-PCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM----------------- | |||||||||||||||||||
| 9 | 5wiuA | 0.23 | 0.23 | 0.84 | 2.73 | Download | ---------------------------------GAAALVGGVLLIGAVLAGNSLVCVSVATERALQTPTNSFIVSLAAADLLLALLVLPLFVYSEVQGAWLLSPRLCDALMAMDVMLCTASIFNLCAISVDRFVAVAVPLRYNRQGGS-RRQLLLIGATWLLSAAVAAPVLCGLN----------DPAVCRL-EDRDYVVYSSVCSFFLPCPLMLLLYWATFRGLQRWEVARRADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPPEMKD---------FRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLAKITGRERKAMRVLPVVVGAFLLCWTPFFVVHITQALCPACSVPPRLVSAVTWLGYVNSALNPVIYTVFNAEFRNVFRKA------------------- | |||||||||||||||||||
| 10 | 5dsgA | 0.26 | 0.27 | 0.88 | 2.27 | Download | ---------------------GPSSHNRYETVEMVFIATVTGSLSLVTVVGNILVMLSIKVNRQLQTVNNYFLFSLACADLIIGAFSMNLYTVYIIKGYWPLGAVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPARRT-TKMAGLMIAAAWVLSFVLWAPAILFWQFVVGKRTVPDNQCFIQFLSNPAVTFGTAIAAFYLPVVIMTVLYIHISLASRSRVNIFEMLRIDEEAEKLFNQDVDAAVRGILRNAKLKPVYDSLDAVRRAALINMVFQMGETGVAGFTN---------------SLRMLQQKRWDEAAVNLAKSRWYNQTPNRAKRVITTFRTGTWDARKVTRTIFAILLAFILTWTPYNVMVLVNTFCQS-CIPDTVWSIGYWLCYVNSTINPACYALCNATFKKTFRHLLLC---------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
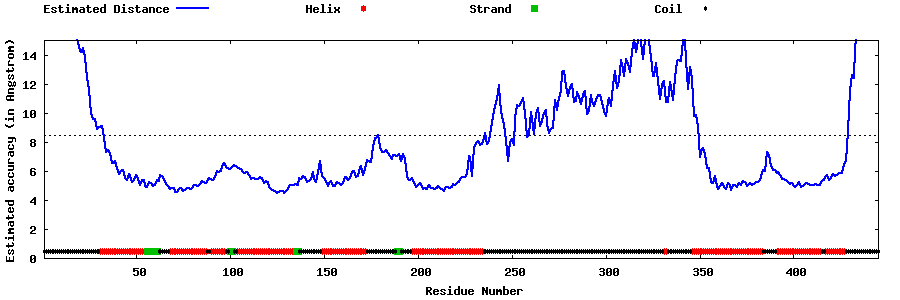
| Generated 3D models | Estimated local accuracy of models | ||
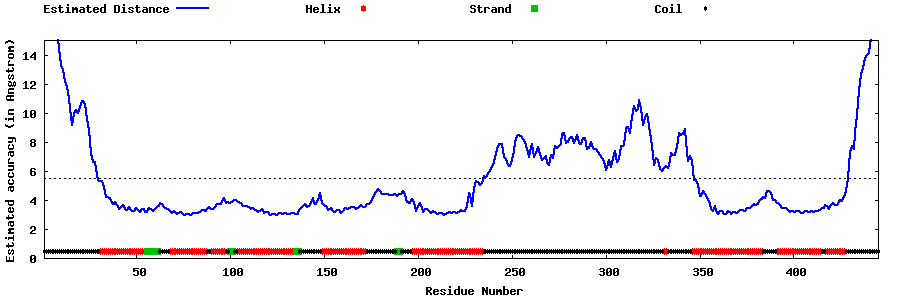 | |||
|
|
|||
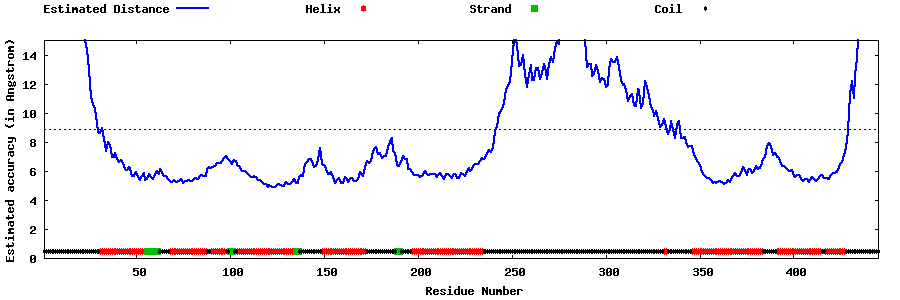 | |||
|
| |||
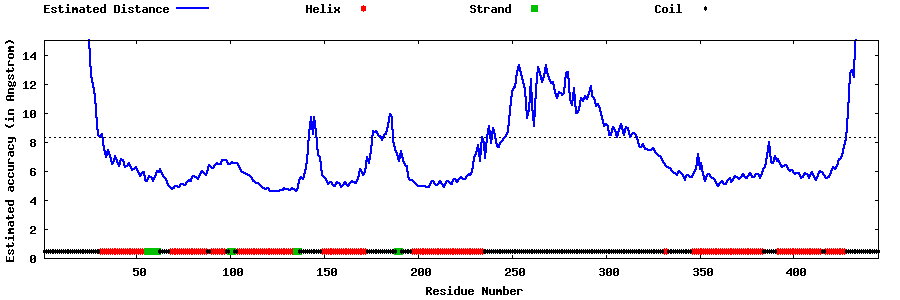 | |||
|
| |||
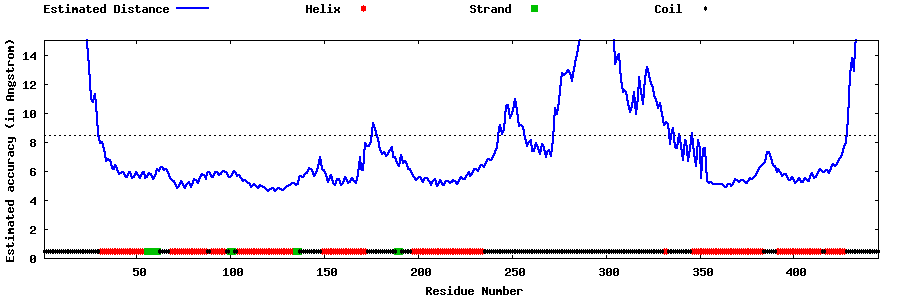 | |||
|
| |||
 | |||
|
| |||