GPCR-I-TASSER results for O43869
[Click on O43869_results.tar.bz2 to download the tarball file including all modeling results listed on this page]
| Submitted Sequence |
| >O43869 MWQEYYFLNVFFPLLKVCCLTINSHVVILLPWECYHLIWKILPYIGTTVGSMEEYNTSST DFTFMGLFNRKETSGLIFAIISIIFFTALMANGVMIFLIQTDLRLHTPMYFLLSHLSLID MMYISTIVPKMLVNYLLDQRTISFVGCTAQHFLYLTLVGAEFFLLGLMAYDRYVAICNPL RYPVLMSRRVCWMIIAGSWFGGSLDGFLLTPITMSFPFCNSREINHFFCEAPAVLKLACA DTALYETVMYVCCVLMLLIPFSVVLASYARILTTVQCMSSVEGRKKAFATCSSHMTVVSL FYGAAMYTYMLPHSYHKPAQDKVLSVFYTILTPMLNPLIYSLRNKDVTGALKRALGRFKG PQRVSGGVF |
| Predicted Secondary Structure |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | MWQEYYFLNVFFPLLKVCCLTINSHVVILLPWECYHLIWKILPYIGTTVGSMEEYNTSSTDFTFMGLFNRKETSGLIFAIISIIFFTALMANGVMIFLIQTDLRLHTPMYFLLSHLSLIDMMYISTIVPKMLVNYLLDQRTISFVGCTAQHFLYLTLVGAEFFLLGLMAYDRYVAICNPLRYPVLMSRRVCWMIIAGSWFGGSLDGFLLTPITMSFPFCNSREINHFFCEAPAVLKLACADTALYETVMYVCCVLMLLIPFSVVLASYARILTTVQCMSSVEGRKKAFATCSSHMTVVSLFYGAAMYTYMLPHSYHKPAQDKVLSVFYTILTPMLNPLIYSLRNKDVTGALKRALGRFKGPQRVSGGVF | |
| CCCCCHHHHHHHHHHHHHCSSCCSSSSSSSSCCHHHHHHHHCCCHHHHHHHHHCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCSSCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCSSSSSHHCHHHHHCCHHHCCCCHHHHHHHHHHHHHCCCHHHCCCCCC | |
| 974204345327888786041321489998600122677633203458888743886313669965899944488999999999999999889997989757775573889987899998753600589999985369968848999999999999999999999999765056305454588415886999999999999999999999999135889989678761482888888713751989999999999999989999999999999981387703253076607878997999987577515288999997669689821740776320254214649999999999985346232046889 |
| Predicted Solvent Accessibility |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | MWQEYYFLNVFFPLLKVCCLTINSHVVILLPWECYHLIWKILPYIGTTVGSMEEYNTSSTDFTFMGLFNRKETSGLIFAIISIIFFTALMANGVMIFLIQTDLRLHTPMYFLLSHLSLIDMMYISTIVPKMLVNYLLDQRTISFVGCTAQHFLYLTLVGAEFFLLGLMAYDRYVAICNPLRYPVLMSRRVCWMIIAGSWFGGSLDGFLLTPITMSFPFCNSREINHFFCEAPAVLKLACADTALYETVMYVCCVLMLLIPFSVVLASYARILTTVQCMSSVEGRKKAFATCSSHMTVVSLFYGAAMYTYMLPHSYHKPAQDKVLSVFYTILTPMLNPLIYSLRNKDVTGALKRALGRFKGPQRVSGGVF | |
| 613421000101120111002121300000203323102200332333153045110200000000003325011100130333233133323200200100030000001002000010010100100200000016521000100100110000001100100010010000000210100020033000000000010002002000000000100342301000003200010001003210110122013313311310330132000000103136123100000000000000112000000010316324434000001102310330030000103101300220033231254146445 | |
| Values range from 0 (buried residue) to 9 (highly exposed residue) | |
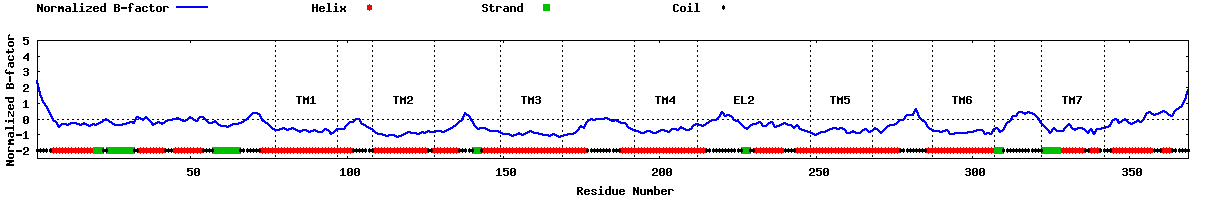
| Predicted normalized B-factor |
| Top 10 templates used by GPCR-I-TASSER |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCHHHHHHHHHHHHHCSSCCSSSSSSSSCCHHHHHHHHCCCHHHHHHHHHCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCSSCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCSSSSSHHCHHHHHCCHHHCCCCHHHHHHHHHHHHHCCCHHHCCCCCC MWQEYYFLNVFFPLLKVCCLTINSHVVILLPWECYHLIWKILPYIGTTVGSMEEYNTSSTDFTFMGLFNRKETSGLIFAIISIIFFTALMANGVMIFLIQTDLRLHTPMYFLLSHLSLIDMMYISTIVPKMLVNYLLDQRTISFVGCTAQHFLYLTLVGAEFFLLGLMAYDRYVAICNPLRYPVLMSRRVCWMIIAGSWFGGSLDGFLLTPITMSFPFCNSREINHFFCEAPAVLKLACADTALYETVMYVCCVLMLLIPFSVVLASYARILTTVQCMSSVEGRKKAFATCSSHMTVVSLFYGAAMYTYMLPHSYHKPAQDKVLSVFYTILTPMLNPLIYSLRNKDVTGALKRALGRFKGPQRVSGGVF | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.13 | 0.18 | 0.94 | 1.61 | Download | --EDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPS---------PSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALLNPVLYAFLDENFKRCFRQLCRKPCG--------- | |||||||||||||||||||
| 2 | 3v2yA | 0.18 | 0.19 | 0.72 | 1.98 | Download | --------------------------------------------------------------------DKENSIKLTSVVFILICCFIILENIFVLLTIWKTKKFHRPMYYFIGNLALSDLLAGVAYTANLLLSGATTYK-LTPAQWFLREGSMFVALSASVFSLLAIAIERYITMLKN--------NFRLFLLISACWVISLILGG--------LPIMGWNCISA--------LSSCSTVLPLYHKHYILFCTTVFTLLLLSIVILYCRIYSLVRTRNSRSSENALLKTVIIVLSVFIACWAPLFILLLLDVGCKVKTCDILFRLVLAVLNSGTNPIIYTLTNKEMRRAFIRIMGR------------ | |||||||||||||||||||
| 3 | 4iaqA | 0.17 | 0.17 | 0.71 | 1.96 | Download | --------------------------------------------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR----WVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL---------PPCVVNTDH-----------------ILYTVYSTVGAFY---FPTLLLIALYGRIYVEARSQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK----------- | |||||||||||||||||||
| 4 | 3emlA | 0.18 | 0.19 | 0.75 | 3.09 | Download | ----------------------------------------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAA-KSLAIIVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ------- | |||||||||||||||||||
| 5 | 2rh1A | 0.20 | 0.17 | 0.76 | 2.59 | Download | ---------------------------------------------------------------------DEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYR----ATHQEAINCYAEET----CCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLNKLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLCL------------ | |||||||||||||||||||
| 6 | 4buoA | 0.21 | 0.18 | 0.76 | 2.11 | Download | -------------------------------------------------------NSD------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKS---TVDYYLGSLALSDLLILLLAMPVELYNFIWVHWAFGDAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMLQNLSGDGTHPGGLVCTP-------IVDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVM---PGRVQALRRGVLVLRVVIAFVVCLMFCYISD-EQWTTFLFDFYHYFYMYVSAAINPILYNLVSANFRQVFLSTL-------------- | |||||||||||||||||||
| 7 | 3emlA | 0.19 | 0.19 | 0.75 | 3.46 | Download | -----------------------------------------------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ------- | |||||||||||||||||||
| 8 | 2lnlA | 0.15 | 0.18 | 0.76 | 2.91 | Download | ----------------------------------------------------------------PCMLEETLNKYVVIIAYALVFLLSLLGNSLVMLVILYSRVGRSVTDVYLLNLALADLLFAL-TLPIWAASKV-NGWIFGTFLCKVVSLLKEVNFYSGILLLACISVDRYLAIVHATRTLTQKR-HLVKFVCLGCWGLSMNLSLPFFLFR---------QAYHPNNSSPVCYEVLGNDTAKWRMLRILPHTFGFIVPLFVMLFCYGFTLRTLFKAH-MGQKHRAMRVIFAVVLIFLLCWLPYNLVLLADTTQVIQNIGRDATEILGFLHSCLNPIIYAFIGQNFRHGFLKILAMHG---------- | |||||||||||||||||||
| 9 | 4buoA | 0.19 | 0.18 | 0.77 | 2.92 | Download | -------------------------------------------------------NSDL------DVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGTPI----------VDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDEQWTNALVYVSAAINPILYNLVSANFRQVFLSTL-------------- | |||||||||||||||||||
| 10 | 3emlA | 0.18 | 0.19 | 0.76 | 4.47 | Download | ----------------------------------------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQSQGCGEGQVAC--------LFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Top 5 Models predicted by GPCR-I-TASSER |
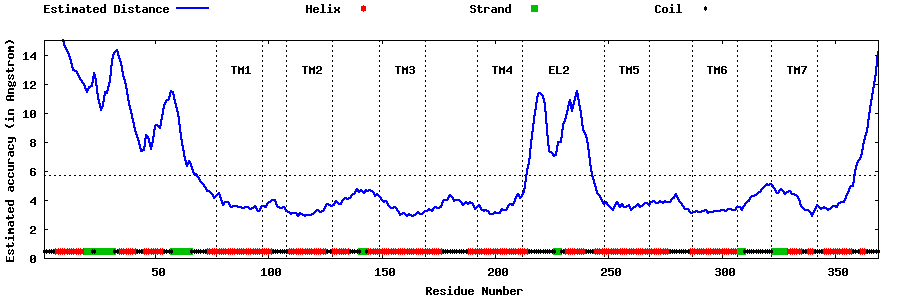
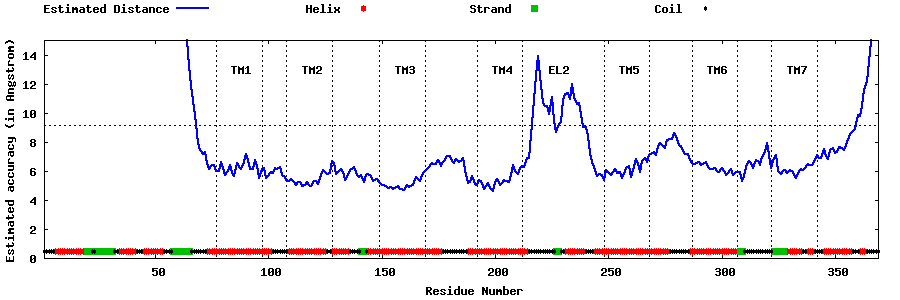
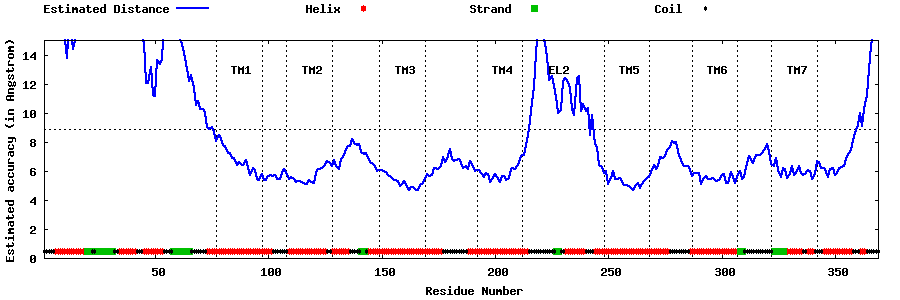
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
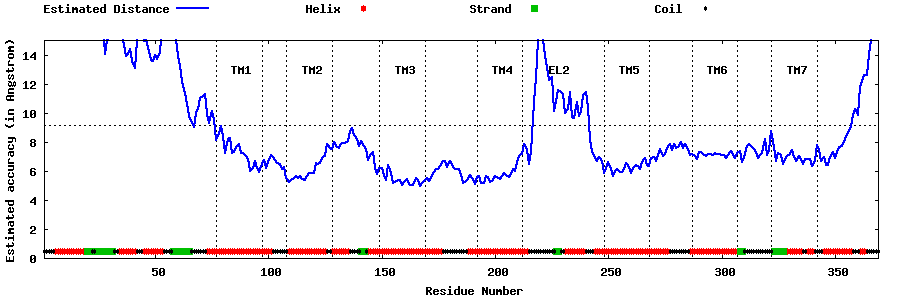 | |||
|
| |||
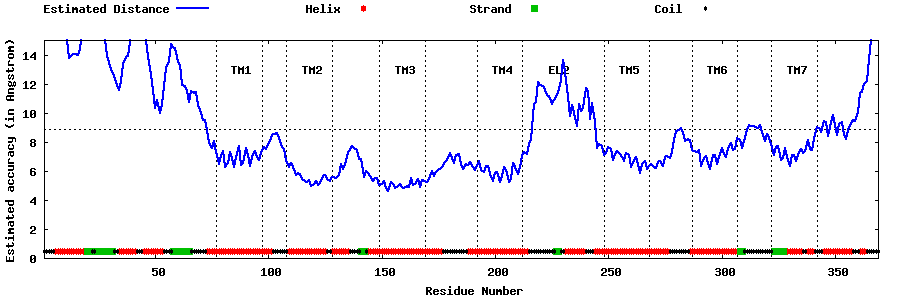 | |||
|
| |||
| Please cite following articles when you use the GPCR-I-TASSER server: | |
| J Zhang, J Yang, R Jang, Y Zhang. Hybrid structure modeling of G protein-coupled receptors in the human genome, submitted (2015). |