GPCR-I-TASSER results for Q5JRS4
[Click on Q5JRS4_results.tar.bz2 to download the tarball file including all modeling results listed on this page]
| Submitted Sequence |
| >Q5JRS4 MPKLNSTFVTEFLFEGFSSFRRQHKLVFFVVFLTLYLLTLSGNVIIMTIIRLDHHLHTPM YFFLCMLSISETCYTVAIIPHMLSGLLNPHQPIATQSCATQLFFYLTFGINNCFLLTVMG YDRYVAICNPLRYSVIMGKRACIQLASGSLGIGLGMAIVQVTSVFGLPFCDAFVISHFFC DVRHLLKLACTDTTVNEIINFVVSVCVLVLPMGLVFISYVLIISTILKIASAEGQKKAFA TCASHLTVVIIHYGCASIIYLKPKSQSSLGQDRLISVTYTHHSPTEPCCVQPEEQGGQRC SAQSRGAKNSVSLMKRGCEGFSFAFINMY |
| Predicted Secondary Structure |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | MPKLNSTFVTEFLFEGFSSFRRQHKLVFFVVFLTLYLLTLSGNVIIMTIIRLDHHLHTPMYFFLCMLSISETCYTVAIIPHMLSGLLNPHQPIATQSCATQLFFYLTFGINNCFLLTVMGYDRYVAICNPLRYSVIMGKRACIQLASGSLGIGLGMAIVQVTSVFGLPFCDAFVISHFFCDVRHLLKLACTDTTVNEIINFVVSVCVLVLPMGLVFISYVLIISTILKIASAEGQKKAFATCASHLTVVIIHYGCASIIYLKPKSQSSLGQDRLISVTYTHHSPTEPCCVQPEEQGGQRCSAQSRGAKNSVSLMKRGCEGFSFAFINMY | |
| CCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCSSSSSCCCHHCCCCHCCCCCCHHHHHHHHHHHHHCCCHHHHHHCCCCHCCSSSSCC | |
| 99988733467778659999808999999999999999999889995878628875574889988899997888721879898985579976868999999999999999999999999754145116555488404776999999999999999999999998263789979668861581898888505873999999999999999859999999999999982287736564254466878997999874254168178999988788199985003221623100104499999999999641021777414430100424429 |
| Predicted Solvent Accessibility |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | MPKLNSTFVTEFLFEGFSSFRRQHKLVFFVVFLTLYLLTLSGNVIIMTIIRLDHHLHTPMYFFLCMLSISETCYTVAIIPHMLSGLLNPHQPIATQSCATQLFFYLTFGINNCFLLTVMGYDRYVAICNPLRYSVIMGKRACIQLASGSLGIGLGMAIVQVTSVFGLPFCDAFVISHFFCDVRHLLKLACTDTTVNEIINFVVSVCVLVLPMGLVFISYVLIISTILKIASAEGQKKAFATCASHLTVVIIHYGCASIIYLKPKSQSSLGQDRLISVTYTHHSPTEPCCVQPEEQGGQRCSAQSRGAKNSVSLMKRGCEGFSFAFINMY | |
| 77542303000000000063352010100230233123113323200200100330000002102000020320110320300010116533010300110110000002100100010000000000210200020033000000010001002103100100010200462301000003200020002203100210112013313312310330222000000103136323100000000100000112000000010317325434000003103310331010001136203300320243320320353032201001234 | |
| Values range from 0 (buried residue) to 9 (highly exposed residue) | |
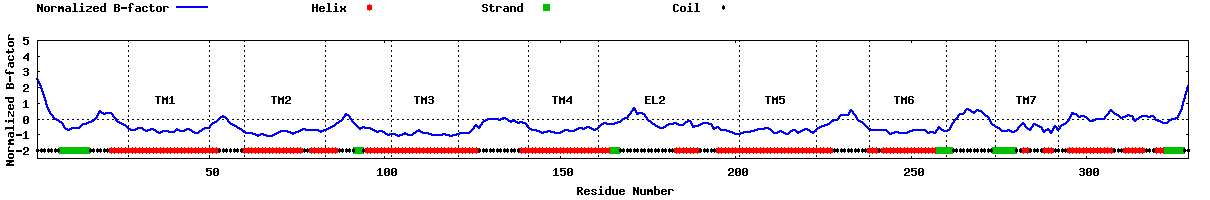
| Predicted normalized B-factor |
| Top 10 templates used by GPCR-I-TASSER |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCSSSSSCCCHHCCCCHCCCCCCHHHHHHHHHHHHHCCCHHHHHHCCCCHCCSSSSCC MPKLNSTFVTEFLFEGFSSFRRQHKLVFFVVFLTLYLLTLSGNVIIMTIIRLDHHLHTPMYFFLCMLSISETCYTVAIIPHMLSGLLNPHQPIATQSCATQLFFYLTFGINNCFLLTVMGYDRYVAICNPLRYSVIMGKRACIQLASGSLGIGLGMAIVQVTSVFGLPFCDAFVISHFFCDVRHLLKLACTDTTVNEIINFVVSVCVLVLPMGLVFISYVLIISTILKIASAEGQKKAFATCASHLTVVIIHYGCASIIYLKPKSQSSLGQDRLISVTYTHHSPTEPCCVQPEEQGGQRCSAQSRGAKNSVSLMKRGCEGFSFAFINMY | |||||||||||||||||||||||||
| 1 | 3v2yA | 0.18 | 0.20 | 0.81 | 1.91 | Download | -----------------SADKENSIKLTSVVFILICCFIILENIFVLLTIWKTKKFHRPMYYFIGNLALSDLLAGVAYTANLLLSGATTYK-LTPAQWFLREGSMFVALSASVFSLLAIAIERYITMLKN--------NFRLFLLISACWVISLILGG--------LPIMGWNCISALSSCS------TVLPLYHKHYILFCTTVFTL--LLLSIVILYCRIYSLVRTRNSRSSENALLKTVIIVLSVFIACWAPLFILLLLDVGCKVKTCDILFRAEYFLVLSGTNPIIYTLTNKEMRRAFIRIMGRP-------------------- | |||||||||||||||||||
| 2 | 4iaqA | 0.11 | 0.21 | 0.81 | 2.24 | Download | -------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTG--RWTLVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL---------PPCVVNT----------------DHILYTVYSTVGAFY----FPTLLLIALYGRIYVEARSRIIQLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK-------------------- | |||||||||||||||||||
| 3 | 3emlA | 0.16 | 0.19 | 0.84 | 3.51 | Download | ---------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ---------------- | |||||||||||||||||||
| 4 | 3uonA | 0.16 | 0.22 | 0.83 | 2.57 | Download | ---------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIV--GVRTVEDGECYIQFF---------SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCCIPNTVWTIGYWLCYINSTNPACYALCNATFKKTFKHLLM----------------------- | |||||||||||||||||||
| 5 | 4bvnA | 0.14 | 0.16 | 0.83 | 2.10 | Download | -------------------LSQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASVETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMH----WWRDEDPQALKCYQDP----GCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKE-----REHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNLVPKWLFVAFNWLGYANSAMNPIILC-RSPDFRKAFKRLLA---------------------- | |||||||||||||||||||
| 6 | 3emlA | 0.16 | 0.19 | 0.84 | 3.95 | Download | ----------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ---------------- | |||||||||||||||||||
| 7 | 4bvnA | 0.13 | 0.16 | 0.84 | 3.08 | Download | -------------------LSQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASVETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWW----RDEDPQALKCYQDP----GCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQ---MREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNLVPKWLFVAFNWLGYANSAMNPIILC-RSPDFRKAFKRLLA---------------------- | |||||||||||||||||||
| 8 | 2rh1A | 0.17 | 0.19 | 0.67 | 2.19 | Download | --------------------DEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQM----HWYRATHQEAINCYAEE----TCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLN-------------------------------------------------------------------------------IFEMLRIDEGLRLKIYKDT | |||||||||||||||||||
| 9 | 3emlA | 0.15 | 0.19 | 0.84 | 5.21 | Download | ---------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ---------------- | |||||||||||||||||||
| 10 | 3emlA | 0.15 | 0.19 | 0.84 | 5.43 | Download | IMG---------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLV-----------PLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWL-PLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR----------------Q | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Top 5 Models predicted by GPCR-I-TASSER |
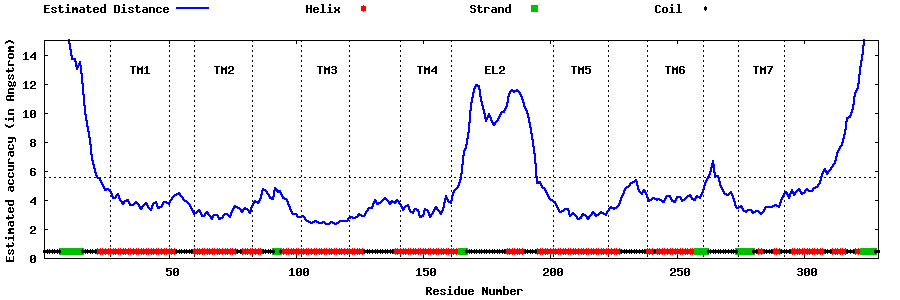
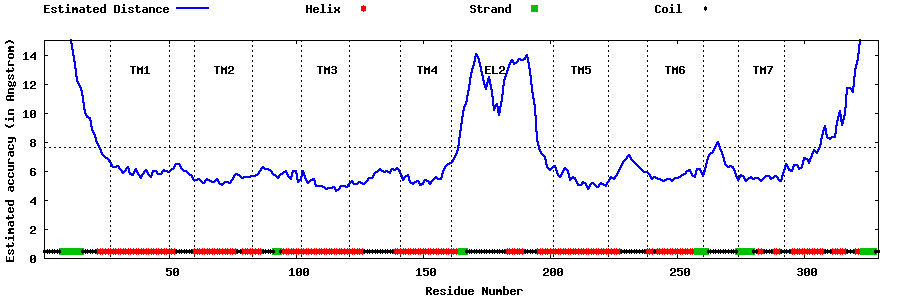
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
| Please cite following articles when you use the GPCR-I-TASSER server: | |
| J Zhang, J Yang, R Jang, Y Zhang. Hybrid structure modeling of G protein-coupled receptors in the human genome, submitted (2015). |