GPCR-I-TASSER results for Q8NGT9
[Click on Q8NGT9_results.tar.bz2 to download the tarball file including all modeling results listed on this page]
| Submitted Sequence |
| >Q8NGT9 MGENQTMVTEFLLLGFLLGPRIQMLLFGLFSLFYIFTLLGNGAILGLISLDSRLHTPMYF FLSHLAVVDIAYTRNTVPQMLANLLHPAKPISFAGCMTQTFLCLSFGHSECLLLVLMSYD RYVAICHPLRYSVIMTWRVCITLAVTSWTCGSLLALAHVVLILRLPFSGPHEINHFFCEI LSVLRLACADTWLNQVVIFAACVFFLVGPPSLVLVSYSHILAAILRIQSGEGRRKAFSTC SSHLCVVGLFFGSAIIMYMAPKSRHPEEQQKVFFLFYSFFNPTLNPLIYSLRNGEVKGAL RRALGKESHS |
| Predicted Secondary Structure |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | MGENQTMVTEFLLLGFLLGPRIQMLLFGLFSLFYIFTLLGNGAILGLISLDSRLHTPMYFFLSHLAVVDIAYTRNTVPQMLANLLHPAKPISFAGCMTQTFLCLSFGHSECLLLVLMSYDRYVAICHPLRYSVIMTWRVCITLAVTSWTCGSLLALAHVVLILRLPFSGPHEINHFFCEILSVLRLACADTWLNQVVIFAACVFFLVGPPSLVLVSYSHILAAILRIQSGEGRRKAFSTCSSHLCVVGLFFGSAIIMYMAPKSRHPEEQQKVFFLFYSFFNPTLNPLIYSLRNGEVKGALRRALGKESHS | |
| CCCCCCSSSSSSSSCCCCCHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCSSSSSHCCHCHCCCCHHHCCCCHHHHHHHHHHHHCCCCC | |
| 9999744346678679998508999999999999999998999999997488756758999878999988887028899999846899658689999999999999999999999997640775065545882238879999999999999999999999991348999894689625858999897136429999999999999998799999999999999822866154651431658689989998742532680789999877780898635051022025533146399999999998462589 |
| Predicted Solvent Accessibility |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | MGENQTMVTEFLLLGFLLGPRIQMLLFGLFSLFYIFTLLGNGAILGLISLDSRLHTPMYFFLSHLAVVDIAYTRNTVPQMLANLLHPAKPISFAGCMTQTFLCLSFGHSECLLLVLMSYDRYVAICHPLRYSVIMTWRVCITLAVTSWTCGSLLALAHVVLILRLPFSGPHEINHFFCEILSVLRLACADTWLNQVVIFAACVFFLVGPPSLVLVSYSHILAAILRIQSGEGRRKAFSTCSSHLCVVGLFFGSAIIMYMAPKSRHPEEQQKVFFLFYSFFNPTLNPLIYSLRNGEVKGALRRALGKESHS | |
| 8662303001000000043151111002301331331233332002001001300000011020000101101003202000001165330102002101100130033002000100200000002102000200330000001200210131031001000002003624010000022100300021031002211231333133123102301220000002032351231001000001000001120000000103163354341000021033203300300101032013002300334538 | |
| Values range from 0 (buried residue) to 9 (highly exposed residue) | |
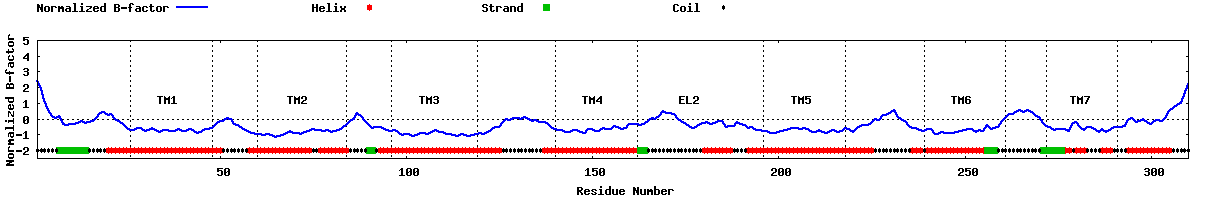
| Predicted normalized B-factor |
| Top 10 templates used by GPCR-I-TASSER |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCSSSSSSSSCCCCCHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCSSSSSHCCHCHCCCCHHHCCCCHHHHHHHHHHHHCCCCC MGENQTMVTEFLLLGFLLGPRIQMLLFGLFSLFYIFTLLGNGAILGLISLDSRLHTPMYFFLSHLAVVDIAYTRNTVPQMLANLLHPAKPISFAGCMTQTFLCLSFGHSECLLLVLMSYDRYVAICHPLRYSVIMTWRVCITLAVTSWTCGSLLALAHVVLILRLPFSGPHEINHFFCEILSVLRLACADTWLNQVVIFAACVFFLVGPPSLVLVSYSHILAAILRIQSGEGRRKAFSTCSSHLCVVGLFFGSAIIMYMAPKSRHPEEQQKVFFLFYSFFNPTLNPLIYSLRNGEVKGALRRALGKESHS | |||||||||||||||||||||||||
| 1 | 4iaqA | 0.18 | 0.20 | 0.86 | 2.31 | Download | -----------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL---------PPCVVNTDH-----------------ILYTVYSTVGAF---YFPTLLLIALYGRIYVEARSRIILLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| 2 | 3emlA | 0.19 | 0.22 | 0.89 | 3.85 | Download | -------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISTG--FCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHI-INCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 3 | 3uonA | 0.19 | 0.23 | 0.89 | 2.56 | Download | -------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIV--GVRTVEDGECYIQFF---------SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM----- | |||||||||||||||||||
| 4 | 4bvnA | 0.19 | 0.21 | 0.88 | 2.06 | Download | ------------------SQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASVETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMM----HWWRDEDPQALKCYQDP----GCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKE-----REHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNLVPKWLFVAFNWLGYANSAMNPIILC-RSPDFRKAFKRLLA----- | |||||||||||||||||||
| 5 | 3emlA | 0.20 | 0.22 | 0.89 | 4.26 | Download | --------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 6 | 4iarA | 0.19 | 0.22 | 0.88 | 2.93 | Download | ----------YIYQD-SISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFW-RQA----------SECVVNT-------D---HILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPI-WFHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| 7 | 4ea3A | 0.19 | 0.23 | 0.86 | 2.98 | Download | ------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLL-TLPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPTSSKAQAVNVAIWALASVVGVPVAIMGSAQ---------VEDEEIDYWGPVFAICIFLFSFIVPVLVISVCYSLMIRRLRGVRLLSGSREKDRNLRRITRLVLVVVAVFVGCWTPVQVFVLAQGL-----GVQPSSETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR--------- | |||||||||||||||||||
| 8 | 3emlA | 0.18 | 0.22 | 0.89 | 5.40 | Download | -------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 9 | 3emlA | 0.19 | 0.22 | 0.89 | 5.62 | Download | I-------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLV-----------PLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWL-PLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 10 | 3emlA | 0.21 | 0.22 | 0.84 | 3.28 | Download | ----------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQSQGCGEGQVACLFEDV--------VPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQ-----LVHAAKSCWLPLHIINCFTFFCPD------CSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKII------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Top 5 Models predicted by GPCR-I-TASSER |
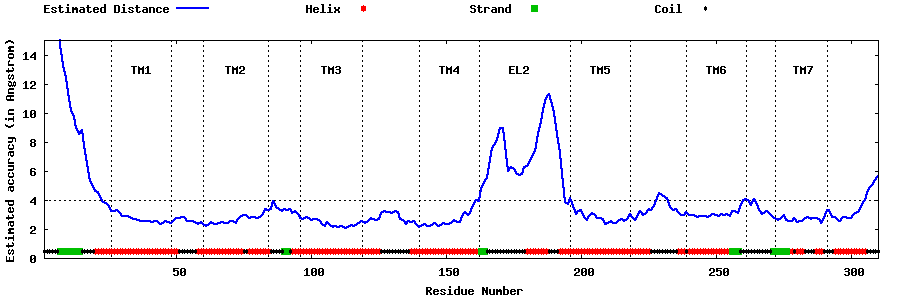
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
| Please cite following articles when you use the GPCR-I-TASSER server: | |
| J Zhang, J Yang, R Jang, Y Zhang. Hybrid structure modeling of G protein-coupled receptors in the human genome, submitted (2015). |