GPCR-I-TASSER results for Q99705
[Click on Q99705_results.tar.bz2 to download the tarball file including all modeling results listed on this page]
| Submitted Sequence |
| >Q99705 MSVGAMKKGVGRAVGLGGGSGCQATEEDPLPNCGACAPGQGGRRWRLPQPAWVEGSSARL WEQATGTGWMDLEASLLPTGPNASNTSDGPDNLTSAGSPPRTGSISYINIIMPSVFGTIC LLGIIGNSTVIFAVVKKSKLHWCNNVPDIFIINLSVVDLLFLLGMPFMIHQLMGNGVWHF GETMCTLITAMDANSQFTSTYILTAMAIDRYLATVHPISSTKFRKPSVATLVICLLWALS FISITPVWLYARLIPFPGGAVGCGIRLPNPDTDLYWFTLYQFFLAFALPFVVITAAYVRI LQRMTSSVAPASQRSIRLRTKRVTRTAIAICLVFFVCWAPYYVLQLTQLSISRPTLTFVY LYNAAISLGYANSCLNPFVYIVLCETFRKRLVLSVKPAAQGQLRAVSNAQTADEERTESK GT |
| Predicted Secondary Structure |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | | | | | | | | | | | | | | | | | | | | | MSVGAMKKGVGRAVGLGGGSGCQATEEDPLPNCGACAPGQGGRRWRLPQPAWVEGSSARLWEQATGTGWMDLEASLLPTGPNASNTSDGPDNLTSAGSPPRTGSISYINIIMPSVFGTICLLGIIGNSTVIFAVVKKSKLHWCNNVPDIFIINLSVVDLLFLLGMPFMIHQLMGNGVWHFGETMCTLITAMDANSQFTSTYILTAMAIDRYLATVHPISSTKFRKPSVATLVICLLWALSFISITPVWLYARLIPFPGGAVGCGIRLPNPDTDLYWFTLYQFFLAFALPFVVITAAYVRILQRMTSSVAPASQRSIRLRTKRVTRTAIAICLVFFVCWAPYYVLQLTQLSISRPTLTFVYLYNAAISLGYANSCLNPFVYIVLCETFRKRLVLSVKPAAQGQLRAVSNAQTADEERTESKGT | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHCHHHHHHHHHHHHHHHHHHHHHHHHCHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCSSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCC | |
| 97663012457788999886666776677888656799865656788985556775310015898887667310237789986657788777766667766442408999899999999999887799760212262568999989999999999999999998299999998389888887688899999999999999999999998588853206424475676888887999999999997999984668927992899624998278999999999999999999999999999999981768986411455541415224689999999997899999999993765248999999999999999997989999982898999999985254047766667788888778848999 |
| Predicted Solvent Accessibility |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | | | | | | | | | | | | | | | | | | | | | MSVGAMKKGVGRAVGLGGGSGCQATEEDPLPNCGACAPGQGGRRWRLPQPAWVEGSSARLWEQATGTGWMDLEASLLPTGPNASNTSDGPDNLTSAGSPPRTGSISYINIIMPSVFGTICLLGIIGNSTVIFAVVKKSKLHWCNNVPDIFIINLSVVDLLFLLGMPFMIHQLMGNGVWHFGETMCTLITAMDANSQFTSTYILTAMAIDRYLATVHPISSTKFRKPSVATLVICLLWALSFISITPVWLYARLIPFPGGAVGCGIRLPNPDTDLYWFTLYQFFLAFALPFVVITAAYVRILQRMTSSVAPASQRSIRLRTKRVTRTAIAICLVFFVCWAPYYVLQLTQLSISRPTLTFVYLYNAAISLGYANSCLNPFVYIVLCETFRKRLVLSVKPAAQGQLRAVSNAQTADEERTESKGT | |
| 64213334125423426444424434746336322321233434242332321442333224333332112232222333332122133332233333144232230000000210120122023232100000000231344300000100220101000000000000000043410001000000100110000000001001002121020100202322221100000000000001000010001202426632100101012461111000001001112110201030001000101313345444444332100000000000010201231000000000123222000001000100002201130001000144104201400221143434534445345454463658 | |
| Values range from 0 (buried residue) to 9 (highly exposed residue) | |
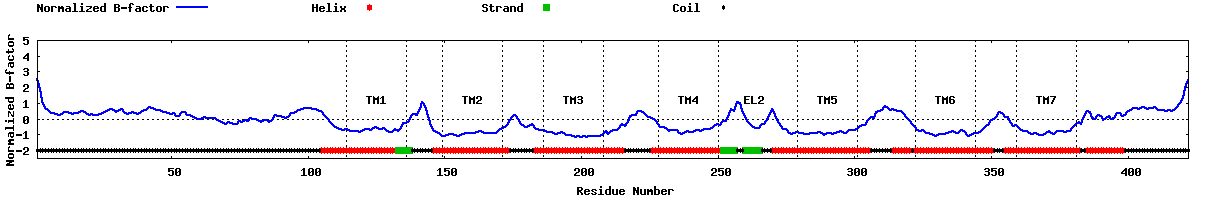
| Predicted normalized B-factor |
| Top 10 templates used by GPCR-I-TASSER |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHCHHHHHHHHHHHHHHHHHHHHHHHHCHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCSSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCC MSVGAMKKGVGRAVGLGGGSGCQATEEDPLPNCGACAPGQGGRRWRLPQPAWVEGSSARLWEQATGTGWMDLEASLLPTGPNASNTSDGPDNLTSAGSPPRTGSISYINIIMPSVFGTICLLGIIGNSTVIFAVVKKSKLHWCNNVPDIFIINLSVVDLLFLLGMPFMIHQLMGNGVWHFGETMCTLITAMDANSQFTSTYILTAMAIDRYLATVHPISSTKFRKPSVATLVICLLWALSFISITPVWLYARLIPFPGGAVGCGIRLPNPDTDLYWFTLYQFFLAFALPFVVITAAYVRILQRMTSSVAPASQRSIRLRTKRVTRTAIAICLVFFVCWAPYYVLQLTQLSISRPTLTFVYLYNAAISLGYANSCLNPFVYIVLCETFRKRLVLSVKPAAQGQLRAVSNAQTADEERTESKGT | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.29 | 0.24 | 0.71 | 4.18 | Download | ------------------------------------------------------------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK---TATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-AVVCMLQFPPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------- | |||||||||||||||||||
| 2 | 4n6hA | 0.25 | 0.29 | 0.93 | 3.48 | Download | VIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAE----QLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK---TATNIYIFNLALADALATSTLPFQSAKYLM-ETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-DGAVVCMLQFPSPWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------- | |||||||||||||||||||
| 3 | 4pxzA | 0.21 | 0.17 | 0.66 | 2.63 | Download | ---------------------------------------------------------------------------------------------SL-----CTRDYKITQVLFPLLYTVLFFVGLITNGLAMRIFFQIRSK----SNFIIFLKNTVISDLLMILTFPFKILSDAKLGTGPLRTFVCQVTSVIFYFTMYISISFLGLITIDRYQKTTRP-----PKNLLGAKILSVVIWAFMFLLSLPNMILTNRQPRDKNVKKCSFLKSEFGLVWHEIVNYICQVIFWINFLIVIVCYTLITKELYRSYVRTADLVGKVPRKKVNVKVFIIIAVFFICFVPFHFARIPYTLSQTAENTLFYVKESTLWLTSLNACLNPFIYFFLCKSF----------------------------------- | |||||||||||||||||||
| 4 | 4n6hA | 0.23 | 0.29 | 0.93 | 4.45 | Download | --WETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK---TATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------- | |||||||||||||||||||
| 5 | 3oduA | 0.25 | 0.23 | 0.72 | 1.90 | Download | -------------------------------------------------------------------------------------------------PCFREENANFNKIFLPTIYSIIFLTGIVGNGLVILVMGYQKKLR---SMTDKYRLHLSVADLLFVITLPFWAVDAV--ANWYFGNFLCKAVHVIYTVNLYSSVWILAFISLDRYLAIVHATNSQRPRKLLAEKVVYVGVWIPALLLTIPDFIFANVSEA-DDRYICDRFYPNDL-WVVVFQFQHIMVGLILPGIVILSCYCIIISKLSHSGSNIFYGSKGHQKRKALKTTVILILAFFACWLPYYIGISIDSFICEFENTVHKWISITEALAFFHCCLNPILYAFLGAKFKTSAQHALTSGRPLEVLFQ---------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.24 | 0.29 | 0.93 | 3.94 | Download | VIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAE----QLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK---TATNIYIFNLALADALATSTLPFQSAKYLM-ETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-DGAVVCMLQFPSSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------- | |||||||||||||||||||
| 7 | 1u19A | 0.20 | 0.19 | 0.81 | 2.74 | Download | ---------------------------------------------------------------MNGT------EGPNFYV-PFSNKTGVVR---SPFEAPQYLAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRT---PLNYILLNLAVADLFMVFGFTTTLY-TSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFR-FGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFG-PIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPL-GDDEASTTVSKTETSQVA- | |||||||||||||||||||
| 8 | 4n6hA | 0.30 | 0.25 | 0.71 | 4.75 | Download | ------------------------------------------------------------------------------------------------GSPGRSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK---TATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPSSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK----PCG------------------ | |||||||||||||||||||
| 9 | 4n6hA | 0.24 | 0.29 | 0.94 | 4.88 | Download | DNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK---TATNIYIFNLALADALATSTLPFQSAKYLM-ETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-DGAVVCMLQFPSPSWWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK----------------------PCG | |||||||||||||||||||
| 10 | 4n6hA | 0.24 | 0.29 | 0.92 | 11.90 | Download | -NLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKM---KTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-DGAVVCMLQFPSPSWWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCR-------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Top 5 Models predicted by GPCR-I-TASSER |
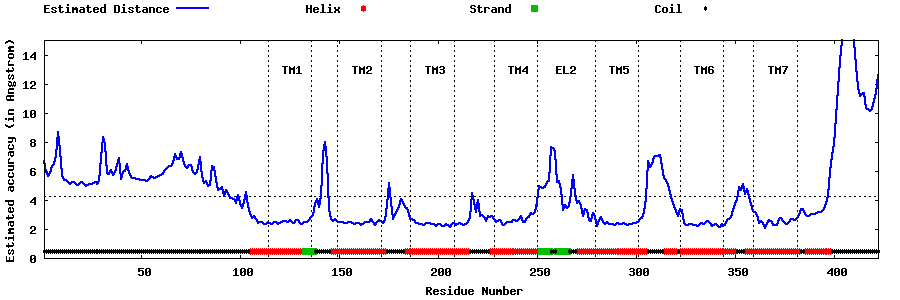
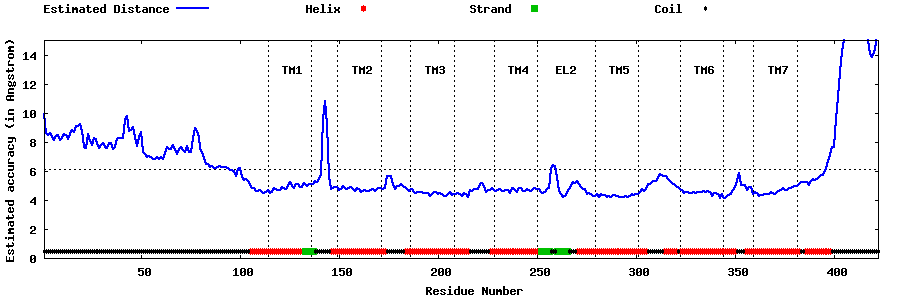
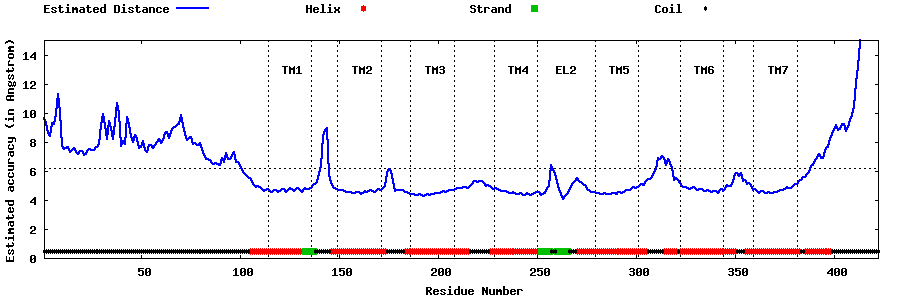
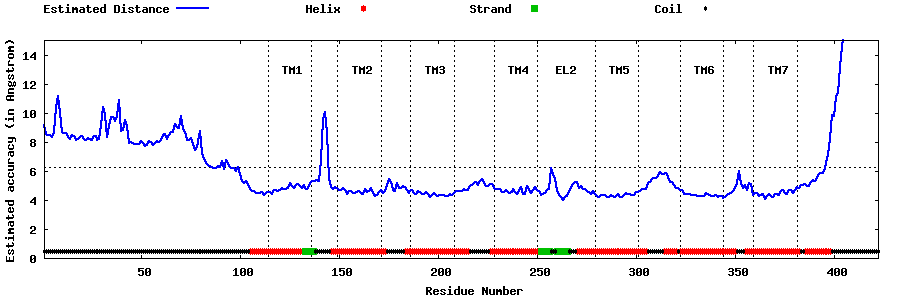
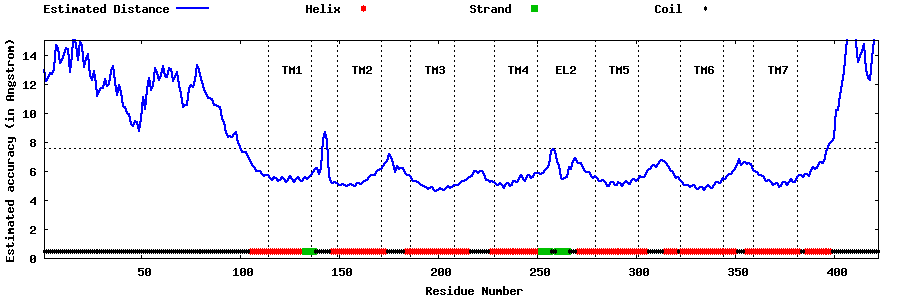
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
| Please cite following articles when you use the GPCR-I-TASSER server: | |
| J Zhang, J Yang, R Jang, Y Zhang. Hybrid structure modeling of G protein-coupled receptors in the human genome, submitted (2015). |