GPCR-I-TASSER results for Q9NQ84
[Click on Q9NQ84_results.tar.bz2 to download the tarball file including all modeling results listed on this page]
| Submitted Sequence |
| >Q9NQ84 MAIHKALVMCLGLPLFLFPGAWAQGHVPPGCSQGLNPLYYNLCDRSGAWGIVLEAVAGAG IVTTFVLTIILVASLPFVQDTKKRSLLGTQVFFLLGTLGLFCLVFACVVKPDFSTCASRR FLFGVLFAICFSCLAAHVFALNFLARKNHGPRGWVIFTVALLLTLVEVIINTEWLIITLV RGSGEGGPQGNSSAGWAVASPCAIANMDFVMALIYVMLLLLGAFLGAWPALCGRYKRWRK HGVFVLLTTATSVAIWVVWIVMYTYGNKQHNSPTWDDPTLAIALAANAWAFVLFYVIPEV SQVTKSSPEQSYQGDMYPTRGVGYETILKEQKGQSMFVENKAFSMDEPVAAKRPVSPYSG YNGQLLTSVYQPTEMALMHKVPSEGAYDIILPRATANSQVMGSANSTLRAEDMYSAQSHQ AATPPKDGKNSQVFRNPYVWD |
| Predicted Secondary Structure |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 | | | | | | | | | | | | | | | | | | | | | | MAIHKALVMCLGLPLFLFPGAWAQGHVPPGCSQGLNPLYYNLCDRSGAWGIVLEAVAGAGIVTTFVLTIILVASLPFVQDTKKRSLLGTQVFFLLGTLGLFCLVFACVVKPDFSTCASRRFLFGVLFAICFSCLAAHVFALNFLARKNHGPRGWVIFTVALLLTLVEVIINTEWLIITLVRGSGEGGPQGNSSAGWAVASPCAIANMDFVMALIYVMLLLLGAFLGAWPALCGRYKRWRKHGVFVLLTTATSVAIWVVWIVMYTYGNKQHNSPTWDDPTLAIALAANAWAFVLFYVIPEVSQVTKSSPEQSYQGDMYPTRGVGYETILKEQKGQSMFVENKAFSMDEPVAAKRPVSPYSGYNGQLLTSVYQPTEMALMHKVPSEGAYDIILPRATANSQVMGSANSTLRAEDMYSAQSHQAATPPKDGKNSQVFRNPYVWD | |
| CCCCCCCCCHHCCSSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCSSSCCCHHHHHHHHHHHHHHHHHHHSSCCCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCCCCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCSHHHHHHHHHHHHHHHHHSSSCCCSSSSSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHCCCCCCCCCCSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 985555430214721214665147989999888777664010263342899999999999999999998751555897178623155067999999999999899852588734346789999999999999999998865541104558642332255689999898751222364246677986434568887089994689838999999999999999999999997056840241189999999999999999998883265434564201146899999999998723740305889824700054013567888766775543356766663655577775434578888777676545666898621542047876777332468778899987778764345333456667789976788760005886679 |
| Predicted Solvent Accessibility |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 | | | | | | | | | | | | | | | | | | | | | | MAIHKALVMCLGLPLFLFPGAWAQGHVPPGCSQGLNPLYYNLCDRSGAWGIVLEAVAGAGIVTTFVLTIILVASLPFVQDTKKRSLLGTQVFFLLGTLGLFCLVFACVVKPDFSTCASRRFLFGVLFAICFSCLAAHVFALNFLARKNHGPRGWVIFTVALLLTLVEVIINTEWLIITLVRGSGEGGPQGNSSAGWAVASPCAIANMDFVMALIYVMLLLLGAFLGAWPALCGRYKRWRKHGVFVLLTTATSVAIWVVWIVMYTYGNKQHNSPTWDDPTLAIALAANAWAFVLFYVIPEVSQVTKSSPEQSYQGDMYPTRGVGYETILKEQKGQSMFVENKAFSMDEPVAAKRPVSPYSGYNGQLLTSVYQPTEMALMHKVPSEGAYDIILPRATANSQVMGSANSTLRAEDMYSAQSHQAATPPKDGKNSQVFRNPYVWD | |
| 733340000000000000303226321137436302320130021420000000000000000001012100110111332232300000000000012012000000021230000000000010101000000000110010022233322100000000101100000000000001223331333434433110000032210000001211120022011000101024136212000000000000000000000000103333333211000000000110322010000010000012336413664333445333534246444343334242234743435444442444333433422231123301443346543423233343454444434433336442334234424356444433323212308 | |
| Values range from 0 (buried residue) to 9 (highly exposed residue) | |
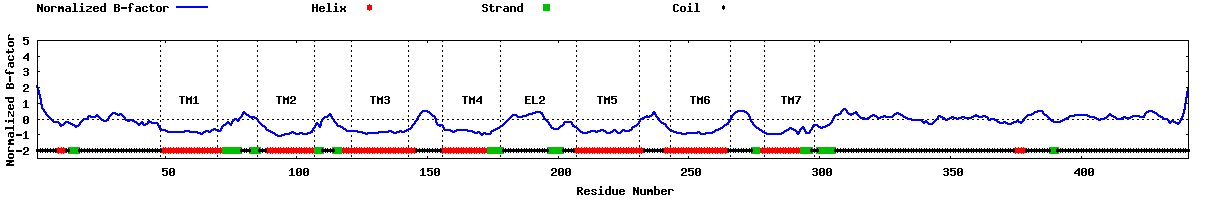
| Predicted normalized B-factor |
| Top 10 templates used by GPCR-I-TASSER |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 | | | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCHHCCSSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCSSSCCCHHHHHHHHHHHHHHHHHHHSSCCCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCCCCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCSHHHHHHHHHHHHHHHHHSSSCCCSSSSSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHCCCCCCCCCCSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MAIHKALVMCLGLPLFLFPGAWAQGHVPPGCSQGLNPLYYNLCDRSGAWGIVLEAVAGAGIVTTFVLTIILVASLPFVQDTKKRSLLGTQVFFLLGTLGLFCLVFACVVKPDFSTCASRRFLFGVLFAICFSCLAAHVFALNFLARKNHGPRGWVIFTVALLLTLVEVIINTEWLIITLVRGSGEGGPQGNSSAGWAVASPCAIANMDFVMALIYVMLLLLGAFLGAWPALCGRYKRWRKHGVFVLLTTATSVAIWVVWIVMYTYGNKQHNSPTWDDPTLAIALAANAWAFVLFYVIPEVSQVTKSSPEQSYQGDMYPTRGVGYETILKEQKGQSMFVENKAFSMDEPVAAKRPVSPYSGYNGQLLTSVYQPTEMALMHKVPSEGAYDIILPRATANSQVMGSANSTLRAEDMYSAQSHQAATPPKDGKNSQVFRNPYVWD | |||||||||||||||||||||||||
| 1 | 4or2A | 0.17 | 0.17 | 0.66 | 2.98 | Download | ---RHGFDILVGQIDDALKLANEGKVTTRNAYIQKYLIPVRYLEWSNIESIIAIAFSCLGILVTLFVTLIFV----LYRDTPVVKSSSRELCYIIGIFLGYVCPFTLIAKPTTTSCYLQRLLVGLSSAMCYSALVTKTNRIARILAGCTRKPRFMQVIIASILISVQLTLVVTLIIMEPPMPILSYPSIKE------VYLICNTSNLGVVAPLGYNGLLIMSCTYYAF--KTRNVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSNYK-------IITTCFAVSLSVTVALGCMFTPKM-YIIIAKPERNV--------------------------------------------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 2 | 3wajA | 0.07 | 0.19 | 0.82 | 1.67 | Download | SVLVLNPWNSVFTWTVRLGGNIHNFPHRIWFDPFTYYPYGSYTHFATSGESLRAVLAFIPAIGGVLAILPVYLLTREVFDRSILGFNDHHIWEAFWQVSALGTFLLAYNRWKGHDAGITIGLYVLSWGAGFIIAPIILAFMFFAFVLAGFVNADRKNLSLVAVVTFAVSALIYLPFAFNYPGFSTI-----------------FYSPFQLLVLLGSAVIAAAFYQIEKWNDVGFFERVGLGRKGMPLAVIVLTALIMAGILYSAYRFLKRRSFPEMALLIWAIAMFSALALSVVF------------------------DKLHFRVAFALLIALAAIYPTYILADAQSSYAGGPNKQWYDALTWMRENTPDGEKYDEYYLQLYPTPQSNKEPFSYPFE-------------------------------------TYGVIA | |||||||||||||||||||
| 3 | 4or2A | 0.19 | 0.14 | 0.56 | 3.87 | Download | ---------------------------------------VRYLEWSNIESIIAIAFSCLGILVTLFVTLIFVLYTPVVKSSSR----ELCYIILAGIFLGYVCPFTLIAKPTTTSCYLQRLLVGLSSAMCYSALVTKTNRIARILA-RKPRSAWAQVIIASILISVQLTLVVTLIIMEPPMPILSYPSIKEV-----YLICNTS-NLGVVAPLGYNGLLIMSCTYYAFKT--RNVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSNYK-------IITTCFAVSLSVTVALGCMFTPKMYIIIA-KPE------------------------------------------------------------------------------------------------------------------------------------ | |||||||||||||||||||
| 4 | 4or2A | 0.18 | 0.16 | 0.66 | 1.63 | Download | KDFRHGFDILVGQIDDALKGKVKEAQAAAEQLKQKYLIPVRYLEWSNIESIIAIAFSCLGILVTLFVTLIFVLYTPVVKSSSRE----LCYIILAGIFLGYVCPFTLIAKPTTTSCYLQRLLVGLSSAMCYSALVTKTNRIARILRKPRFMSAWAQVIIASILISVQLTLVVTLIIMEPPMPILSYPSIKE------VYLICNTSNLGVVAPLGYNGLLIMSCTYYAFKTR--NVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSN-------YKIITTCFAVSLSVTVALGCMFTPK-MYIIIAKPE------------------------------------------------------------------------------------------------------------------------------------ | |||||||||||||||||||
| 5 | 4or2A | 0.19 | 0.16 | 0.65 | 8.45 | Download | DKIDDALKLANEGKVKEAQAAEQLKTTRNAYIQK-YLIPVRYLEWSNIESIIAIAFSCLGILVTLFVTLIFVLDTPVVK----SSSRELCYIILAGIFLGYVCPFTLIAKPTTTSCYLQRLLVGLSSAMCYSALVTKTNRIARILA---FMSAWAQVIIASILISVQLTLVVTLIIMEPPMPILSYP------SIKEVYLICNTSNLGVVAPLGYNGLLIMSCTYYAFKTR--NVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSNY-------KIITTCFAVSLSVTVALGCMFTPKMYIIIAK-PE------------------------------------------------------------------------------------------------------------------------------------ | |||||||||||||||||||
| 6 | 4or2A | 0.17 | 0.16 | 0.65 | 6.32 | Download | QKKLEDKSPEMKDLKLANEGKVKEAEQLKTTRNAYIQKPVRYLEWSNIESIIAIAFSCLGILVTLFVTLIFVL----YRDTPVVKSSSRELCYLAGIFLGYVCPFTLIAKPTTTSCYLQRLLVGLSSAMCYSALVTKTNRIARILA----MSAWAQVIIASILISVQLTLVVTLIIMEPPMPILSYP-----SIKEVYLICNTS-NLGVVAPLGYNGLLIMSCTYYAF--KTRNVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSNYK-------IITTCFAVSLSVTVALGCMFTPK-MYIIIAKPE------------------------------------------------------------------------------------------------------------------------------------ | |||||||||||||||||||
| 7 | 4or2A | 0.18 | 0.16 | 0.66 | 2.74 | Download | KDFRHGFDILVGQIDDALKLANEGLKTTRNAYIQKYLIPVRYLEWSNIESIIAIAFSCLGILVTLFVTLIFVLYTPVVKSSS----RELCYIILAGIFLGYVCPFTLIAKPTTTSCYLQRLLVGLSSAMCYSALVTKTNRIARILRKPRFMSAWAQVIIASILISVQLTLVVTLIIMEPPMPILSYPSIKE------VYLICNTSNLGVVAPLGYNGLLIMSCTYYAFKTR--NVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSN-------YKIITTCFAVSLSVTVALGCMFTPK-MYIIIAKPE------------------------------------------------------------------------------------------------------------------------------------ | |||||||||||||||||||
| 8 | 4or2A | 0.18 | 0.16 | 0.65 | 8.79 | Download | DILGQIDDALKLANEGKVQAAAEQKTTRNAYIQKY-LIPVRYLEWSNIESIIAIAFSCLGILVTLFVTLIFVRDTPVVKSSSRE----LCYIILAGIFLGYVCPFTLIAKPTTTSCYLQRLLVGLSSAMCYSALVTKTNRIARILA---FMSAWAQVIIASILISVQLTLVVTLIIMEPPMP------ILSYPSIKEVYLICNTSNLGVVAPLGYNGLLIMSCTYYAF--KTRNVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSNY-------KIITTCFAVSLSVTVALGCMFTPKMYIIIA-KPE------------------------------------------------------------------------------------------------------------------------------------ | |||||||||||||||||||
| 9 | 4or2A | 0.20 | 0.15 | 0.56 | 2.58 | Download | -------------------------------------IPVRYLEWSNIESIIAIAFSCLGILVTLFVTLIFVLY----RDTVKSSSRELCYIILAGIFLGYVCPFTLIAKPTTTSCYLQRLLVGLSSAMCYSALVTKTNRIARILARKRFMSAWAQVIIASILISVQLTLVVTLIIMEPPMYPSIKE---------VYLICN-TSNLGVVAPLGYNGLLIMSCTYYAFKT--RNVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSNY-------KIITTCFAVSLSVTVALGCMFTPKMYIIIA-KPE------------------------------------------------------------------------------------------------------------------------------------ | |||||||||||||||||||
| 10 | 4or2A | 0.20 | 0.14 | 0.56 | 2.07 | Download | ---------------------------------------VRYLEWSNIESIIAIAFSCLGILVTLFVTLIFVLYTPVVKSSSRE----LCYIILAGIFLGYVCPFTLIAKPTTTSCYLQRLLVGLSSAMCYSALVTKTNRIARILRKPRFMSAWAQVIIASILISVQLTLVVTLIIMEPPMPILSYPSIKE------VYLICNTSNLGVVAPLGYNGLLIMSCTYYAFKT---NVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSNYK-------IITTCFAVSLSVTVALGCMFTPK-MYIIIAKPE------------------------------------------------------------------------------------------------------------------------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Top 5 Models predicted by GPCR-I-TASSER |
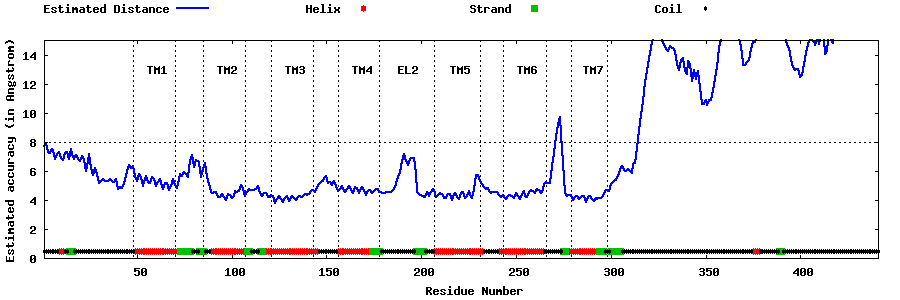
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
Estimated acuracy of domain 1 (1-331) Estimated acuracy of domain 2 (332-441) |
|||
| Please cite following articles when you use the GPCR-I-TASSER server: | |
| J Zhang, J Yang, R Jang, Y Zhang. Hybrid structure modeling of G protein-coupled receptors in the human genome, submitted (2015). |