Welcome to Xi Zhang's Homepage!
Xi Zhang, PhD
Postdoctoral Research Fellow
Department of Computational Medicine and Bioinformatics
Medical School, University of Michigan
100 Washtenaw Avenue
Ann Arbor, MI 48109‐2218, USA
Tel: (734)‐773-2530
Email: xizha@umich.edu
|
Research Interests
Predict protein structure from cryo-EM density map and related field, Computational Biology, Bioinformatics, De Novo Protein structure prediction, Biophysics, Molecular
Modeling, Deep Learning, Algorithm design.
In the last decade, theoretical and technological innovations have been made in the field of cryo-EM, such as single particle analysis and direct electron detection cameras. However, cryo-EM has still been limited to 3-4 Å resolution, and it is challenging to determine sub-2Å structures. Although multiple methods exist such as Flex-EM, RosettaCM2 and EMD-Ref, most of the approaches construct models by optimally refining configurations with density map, which is helpful in atomic model reproduction but cannot improve cryo-EM data resolution. Moreover, these methods are highly relied on the initial predicted conformation. Currently, Dr. Xi Zhang is dedicated to developing state-of-the-art algorithms for high-resolution protein structure De Novo prediction with/from cryo-EM density map. As shown in Figure 1, Dr. Zhang and cooperators develop CRI-TASSER (cryo-EM I-TASSER), a new pipeline integrating the information of cryo-EM density map into the powerful approach I-TASSER for protein structure determination under cryo-EM experimental data. The new approach starts with protein sequence and the corresponding cryo-EM density map, and predicts the 3D structure of the query sequence.
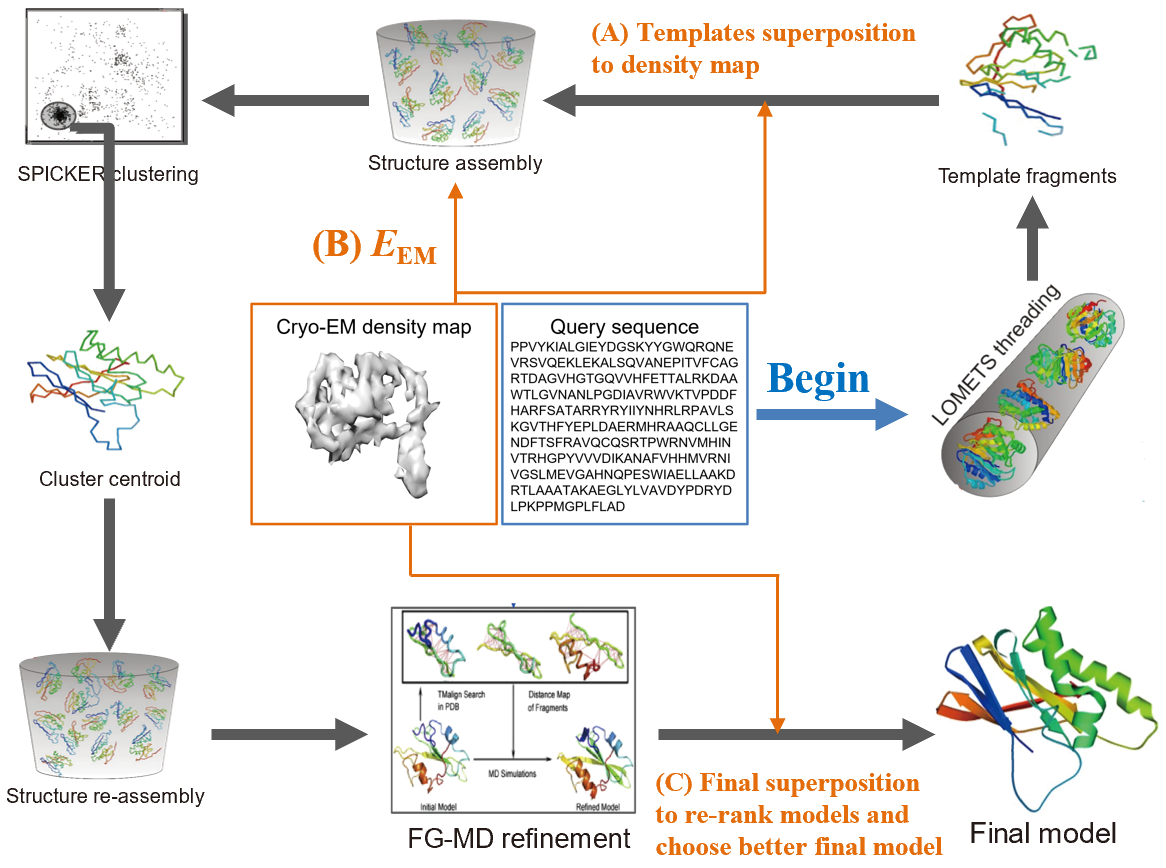
Figure 1. CRI-TASSER pipeline for de novo protein structure determination with inputs of query sequence (blue inset) and the corresponding cryo-EM density map (orange inset).
Education
-
2018.5 Deep Learning Specialization on coursera.org (included 5 courses with certificates)
-
2017.7 Ph.D., School of Physics and Technology, Wuhan University, Wuhan, China
Dissertation: Interaction between ions and polyelectrolytes: From macroions to nucleic acids. Advisor: Prof. Zhi-Jie Tan.
-
2012.9-2017.7 Ph.D. Candidate in Biophysics, School of Physics and Technology, Wuhan University, Wuhan, China
-
2012.7 B.S., School of Physics and Technology, Wuhan University, Wuhan, China
Thesis: Poisson-Boltzmann theory for nucleic acid-ion interactions. Advisor: Prof. Zhi-Jie Tan.
-
2010.3-2012.7 B.S. student in National Scientific Base for Talented Persons in Physics, School of Physics and Technology, Wuhan University, Wuhan, China
-
2009.3-2010.2 B.S. student in Physics, School of Physics and Technology, Wuhan University, Wuhan, China
-
2008.9-2009.2 B.S. student in Software Engineer, International School of Software, Wuhan University, Wuhan, China
Publications
-
Xi Zhang, Biao Zhang, and Yang Zhang*, "Zhang-Server-EM: A meta-server for predicting protein structure from cryo-EM density map" 2019 (in preparation)
-
Xi Zhang, Yang Li, and Yang Zhang*, "REM: De Novo protein structure prediction from cryo-EM density map using deep residual neural network" 2019 (in preparation)
-
Xi Zhang, Biao Zhang, and Yang Zhang*, "CRI-TASSER: De Novo approach for high-resolution protein structure determination with cryo-EM density map" 2019 (in preparation)
-
Biao Zhang, Xi Zhang, Hongbin Shen, and Yang Zhang*, "Refinemental predicted strcture using cryo-EM density map" 2019 (in preparation)
-
Cheng Lin, Xi Zhang, Xiaowei Qiang, Jin-Si Zhang, and Zhi-Jie Tan*. "Apparent repulsion between equally and oppositely charged spherical polyelectrolytes in symmetrical salt solutions." The Journal of Chemical Physics 151(11):114902, 2019. (co-first author)
-
Ju-Hui Liu, Kun Xi, Xi Zhang, Lei Bao, Xinghua Zhang*, and Zhi-Jie Tan*. "Structural Flexibility of DNA-RNA Hybrid Duplex: Stretching and Twist-Stretch Coupling" Biophysical journal, 117:74-86, 2019.
-
Gui Xiong, Kun Xi, Xi Zhang*, and Zhi-Jie Tan*, "To what extent of ion neutralization, multivalent ion distributions around RNA-like macroions can be described by the Poisson-Boltzmann theory?" Chinese Physics B 27:018203, 2018. (co-corresponding author)
-
Jin-Si Zhang, Xi Zhang, Zhong-Liang Zhang, and Zhi-Jie Tan*, "Potential of mean force between oppositely charged nanoparticles: A comprehensive comparison between Poisson-Boltzmann theory and Monte Carlo simulations" Scientific Reports 7, 14145, 2017. (co-first author)
-
Xi Zhang, Lei Bao, Yuan-Yan Wu, Xiao-Long Zhu, and Zhi-Jie Tan*, "Radial distribution function of semiflexible oligomers with stretching flexibility" The Journal of Chemical Physics 147(5):054901, 2017. (featured article; editor recommended as newsworthy)
-
Lei Bao, Xi Zhang, Ya-Zhou Shi, Yuan-Yan Wu, Zhi-Jie Tan*, "Understanding the relative flexibility of RNA and DNA duplexes: stretching and twist-stretch coupling" Biophysical Journal, 112:1094-1104, 2017. (co-first author)
-
Xi Zhang, Jin-Si Zhang, Ya-Zhou Shi, Xiao-Long Zhu, Zhi-Jie Tan*, "Potential of mean force between like-charged nanoparticles: Many-body effect" Scientific Reports 6, 23434, 2016.
-
Lei Bao, Xi Zhang, Lei Jin, Zhi-Jie Tan*, "Flexibility of nucleic acids: from DNA to RNA" Chinese Physics B, 25:018703, 2016
-
Yuan-Yan Wu, Lei Bao, Xi Zhang, Zhi-Jie Tan*, "Flexibility of short DNA helices with finite-length effect: from base pairs to tens of base pairs" The Journal of Chemical Physics, 142:125103, 2015
Presentations at Academic Conference
-
2019.9 Poster presentation, 2019 DCMB Annual Retreat. Oregon, OH, USA.
-
2017.7 Poster presentation, 4th National Conference on Statistical Physics and Complex System. Xi’an, Shanxi, China. (Recognized as Excellent Poster award)
-
2015.7 Poster presentation, 3rd National Conference on Statistical Physics and Complex System. Lanzhou, Gansu, China
-
2014.6 Poster presentation, 8th IUPAP International Conference on Biological Physics. Beijing, China
-
2013.9 Oral presentation, workshop entitled "Computational challenges on complex charge system". Shanghai, China
-
2013.7 Oral presentation, 2nd National Conference on Statistical Physics and Complex System. Qufu, Shandong, China
Hobbies
-
Dr. Xi Zhang loves playing badminton and board games such as chess and Go.



![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218