

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 | |
| | | | | | | | | | | | | | | | | | | | | | | | |
| MASPSLPGSDCSQIIDHSHVPEFEVATWIKITLILVYLIIFVMGLLGNSATIRVTQVLQKKGYLQKEVTDHMVSLACSDILVFLIGMPMEFYSIIWNPLTTSSYTLSCKLHTFLFEACSYATLLHVLTLSFERYIAICHPFRYKAVSGPCQVKLLIGFVWVTSALVALPLLFAMGTEYPLVNVPSHRGLTCNRSSTRHHEQPETSNMSICTNLSSRWTVFQSSIFGAFVVYLVVLLSVAFMCWNMMQVLMKSQKGSLAGGTRPPQLRKSESEESRTARRQTIIFLRLIVVTLAVCWMPNQIRRIMAAAKPKHDWTRSYFRAYMILLPFSETFFYLSSVINPLLYTVSSQQFRRVFVQVLCCRLSLQHANHEKRLRVHAHSTTDSARFVQRPLLFASRRQSSARRTEKIFLSTFQSEAEPQSKSQSLSLESLEPNSGAKPANSAAENGFQEHEV | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 999999998877777777788666632999999999999999989977755058999922899982878999999999999999999999999996689878887927179999999999999999999998682300206386341388888864799999999999999986067413456655564235666534566777752798515737889999987764344689999999999999999998526667777752000046778888778899989999999999968999999999965434578126899999999999999999999899999759999999999848767887767666756767787888874556766678877876777767767766656778888777766778789999999777786554779 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 | |
| | | | | | | | | | | | | | | | | | | | | | | | |
| MASPSLPGSDCSQIIDHSHVPEFEVATWIKITLILVYLIIFVMGLLGNSATIRVTQVLQKKGYLQKEVTDHMVSLACSDILVFLIGMPMEFYSIIWNPLTTSSYTLSCKLHTFLFEACSYATLLHVLTLSFERYIAICHPFRYKAVSGPCQVKLLIGFVWVTSALVALPLLFAMGTEYPLVNVPSHRGLTCNRSSTRHHEQPETSNMSICTNLSSRWTVFQSSIFGAFVVYLVVLLSVAFMCWNMMQVLMKSQKGSLAGGTRPPQLRKSESEESRTARRQTIIFLRLIVVTLAVCWMPNQIRRIMAAAKPKHDWTRSYFRAYMILLPFSETFFYLSSVINPLLYTVSSQQFRRVFVQVLCCRLSLQHANHEKRLRVHAHSTTDSARFVQRPLLFASRRQSSARRTEKIFLSTFQSEAEPQSKSQSLSLESLEPNSGAKPANSAAENGFQEHEV | |
| 761442333332232333313313101001000000001001102310200010000001244021000030013121210001011201000001332000011000100000010000000102000000021000100102321233000000000001000001100000002233232443332223331233333242210000121233221210000011331113000000000000001022334434454444434444344434311100000000000000002110010000000334324310110000010100110020000000000000530040013000011324434444433443433433333332323334444344443442414234454565644543436334444644345333433335348 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 | | | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MASPSLPGSDCSQIIDHSHVPEFEVATWIKITLILVYLIIFVMGLLGNSATIRVTQVLQKKGYLQKEVTDHMVSLACSDILVFLIGMPMEFYSIIWNPLTTSSYTLSCKLHTFLFEACSYATLLHVLTLSFERYIAICHPFRYKAVSGPCQVKLLIGFVWVTSALVALPLLFAMGTEYPLVNVPSHRGLTCNRSSTRHHEQPETSNMSICTNLSSRWTVFQSSIFGAFVVYLVVLLSVAFMCWNMMQVLMKSQKGSLAGGTRPPQLRKSESEESRTARRQTIIFLRLIVVTLAVCWMPNQIRRIMAAAKPKHDWTRSYFRAYMILLPFSETFFYLSSVINPLLYTVSSQQFRRVFVQVLCCRLSLQHANHEKRLRVHAHSTTDSARFVQRPLLFASRRQSSARRTEKIFLSTFQSEAEPQSKSQSLSLESLEPNSGAKPANSAAENGFQEHEV | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.20 | 0.18 | 0.66 | 2.41 | Download | -----------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVR---YTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPF--GELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-------------------------GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS------------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD----PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------------------------------------------------------------------------------- | |||||||||||||||||||
| 2 | 4xeeA | 0.25 | 0.28 | 0.87 | 3.56 | Download | ---------------GPNSDLDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLA-------RKSTVHYHLGSLALSDLLILLLAMPVELYNFIWVHHPWAFGDAGCRGYYFLRDACTYATALNVASLSVERYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNRSADGTHPGG--------------------LVCTPIVDTATVKVVIQVNTFMSFLFPMLVISILNTVIANKLTVMVP-----------------GRVQALRHGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDEQWTTFLFDFYHYFYMLTNALFYASSAINPILYNLVSANFRQVFLSTLNIFEMLRIDEG-LRLKIYKDTEGYYTIGIGHLLTKSPSLNAAKSELDKAIGRNTNGVITKDEAEKLFNQDVDAAVRGILRNAKLKPVYDSLDAV | |||||||||||||||||||
| 3 | 5gliA | 0.21 | 0.20 | 0.71 | 4.00 | Download | -------------ISPPPCQGPIEIKETFKYINTVVSCLVFVLGIIGNSTLL---YIIYKNKCMRNGPNILIASLALGDLLHIVIAIPINVYKLLAEDWPF--GAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWSRIKGIGVPKWTAVEIVLIWVVSVVLAVPEAIGFDIITMDY---------------------KGSYLRICLLHPVMQFYATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLRIDEGGGSGGDRTGTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQNDPNRLLSFLLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSALCC-------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 4 | 4xeeA | 0.28 | 0.28 | 0.87 | 2.15 | Download | -----GPNSDLDVNTD----------IYSKVLVTAIYLALFVVGTVGNSVTLFTL-------ARKSTVHYHLGSLALSDLLILLLAMPVELYNFIWVHHPWAFGDAGCRGYYFLRDACTYATALNVASLSVERYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNRSADGTHPGG-------------------LVCTPIVDTATVKVVIQVNTFMS-FLFPMLVISILNTVIANKLTVMVP-----------------GRVQALRHGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDEQWTTFLFDFYHYFYMLTNALFYASSAINPILYNLVSANFRQVFLSTLNIFRIDEGLRLKIYKDTEGYYTIGIGHLLTKSPSLNAAKSDKAIGRNTNGVITKDEAEKLFNQDVDAAVRGILRNAKLKPVYDSLDAVMGETGV | |||||||||||||||||||
| 5 | 4n6hA | 0.20 | 0.18 | 0.67 | 3.14 | Download | -----------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY---TKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWP--FGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD------GAVVCMLQFPS-----------------PSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLS------------GSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDP----LVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------------------------------------------------------------------------------- | |||||||||||||||||||
| 6 | 2ziy | 0.18 | 0.21 | 0.81 | 1.55 | Download | LRDNETWWYNPSI-IVHPHWEFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKS---LQTPANMFIINLAFSDFTFSLVNFPLMTISCF--LKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLE------------------------GVLCNCSFDYIDSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNAKELRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWV-------TPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETE-DDKDAETE------IPAGESSDAAP--------------------------SADAAQMKE------------------ | |||||||||||||||||||
| 7 | 4djh | 0.21 | 0.21 | 0.66 | 1.17 | Download | -------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVI---IRYTKMKTATNIYIFNLALADALVT-TTMPFQSTVYLMNSWPFG--DVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRED-----------------------VDVIECSLQFPDDWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNINTNGVKQTPNYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA----A------LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP-------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.20 | 0.18 | 0.66 | 2.94 | Download | -------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFG---IVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWP--FGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-------------------------GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLL------------SGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD----PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------------------------------------------------------------------------------- | |||||||||||||||||||
| 9 | 3uon | 0.16 | 0.21 | 0.65 | 1.76 | Download | -----------------------------VVFIVLVAGSLSLVTIIGNILVMVSI---KVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYW--PLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVE---DGECYIQFF-----------------SNA--AV--TFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEGGHAANVTTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPC--------IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM--------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 10 | 4n6hA | 0.20 | 0.18 | 0.66 | 4.21 | Download | ----------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMF---GIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFG--ELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-------------------------GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS------------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD----PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------------------------------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
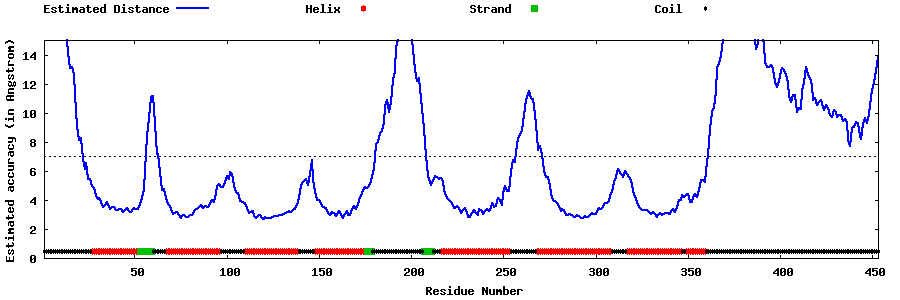 | |||
|
|
|||
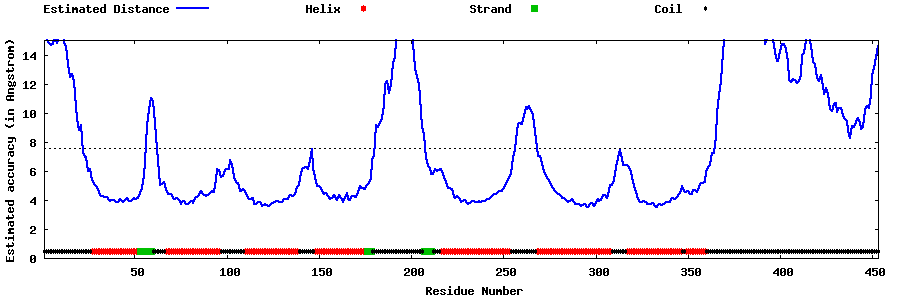 | |||
|
| |||
 | |||
|
| |||
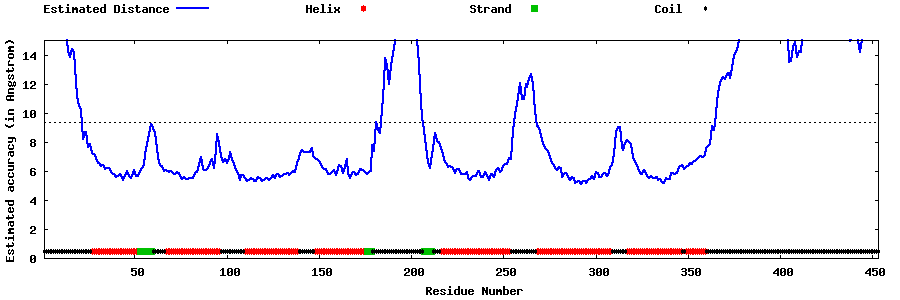 | |||
|
| |||
 | |||
|
| |||