


| HG ID | UniProt ID | Length | C-score | Estimated TM-score |
Estimated RMSD |
Top 5 models |
| HG0170 | Q8TBK4 | 359 | -0.52 | 0.65 ± 0.13 | 7.7 ± 4.3 | 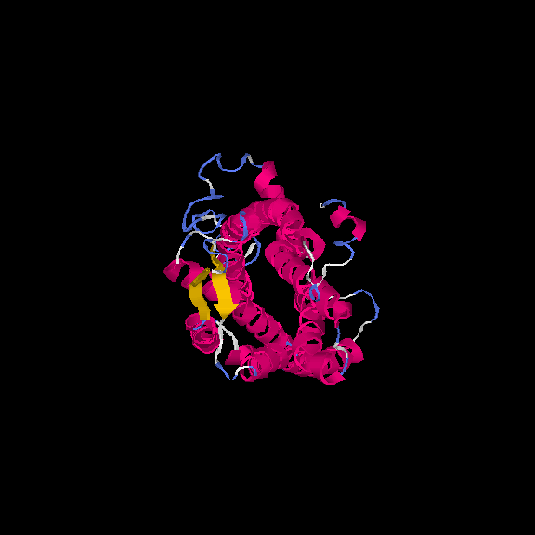 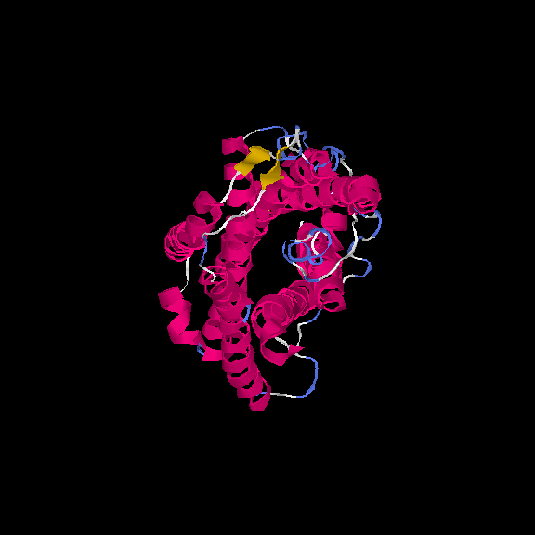 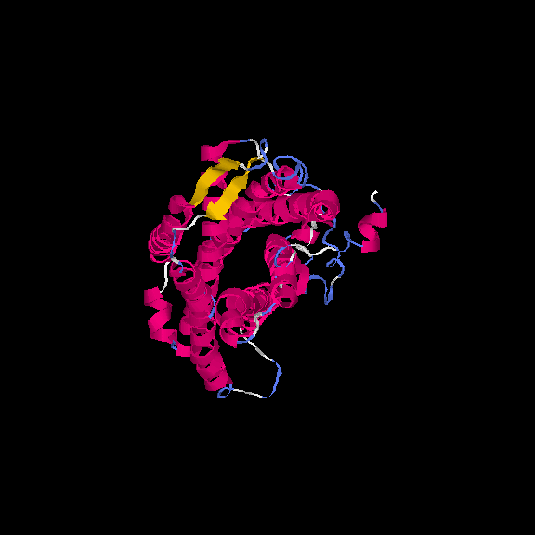 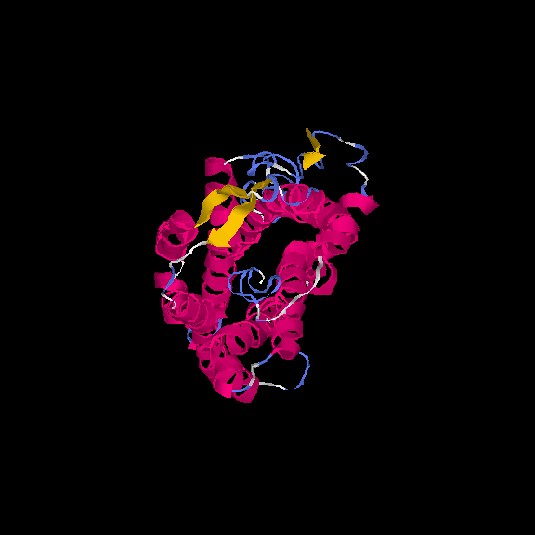 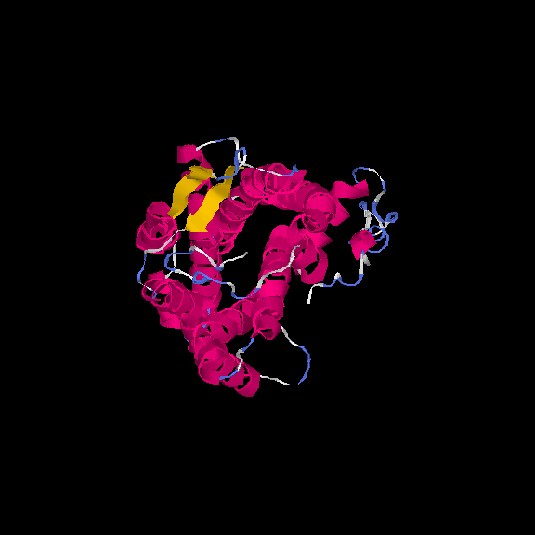 |
| HG0171 | O76002 | 312 | -0.11 | 0.7 ± 0.12 | 6.5 ± 3.9 | 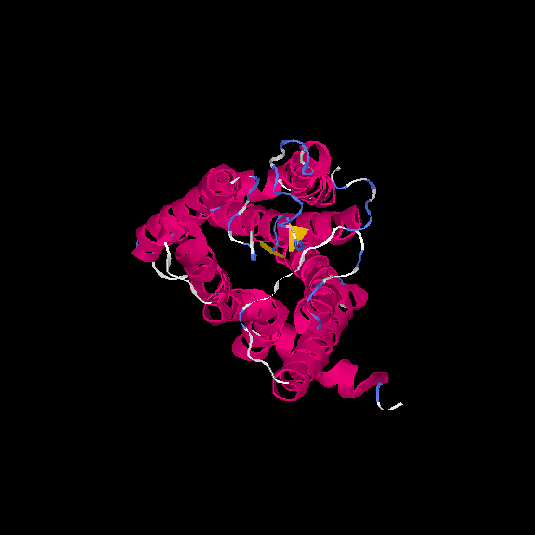 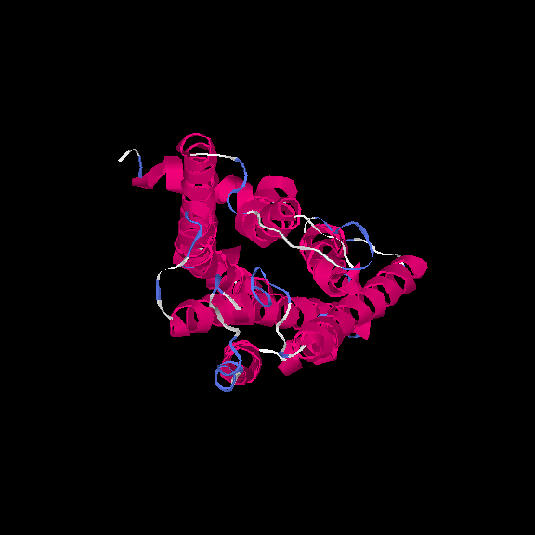 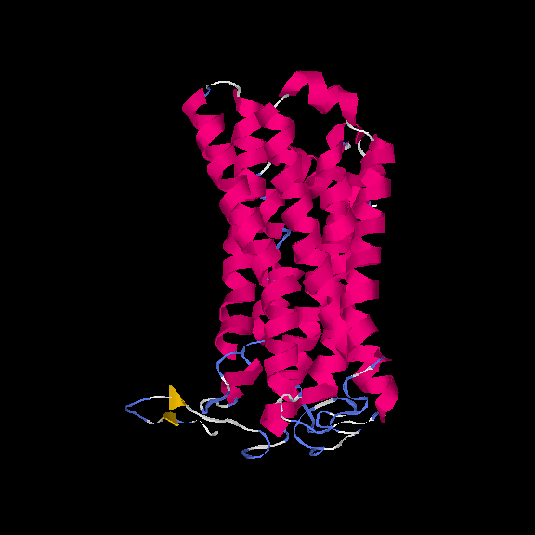 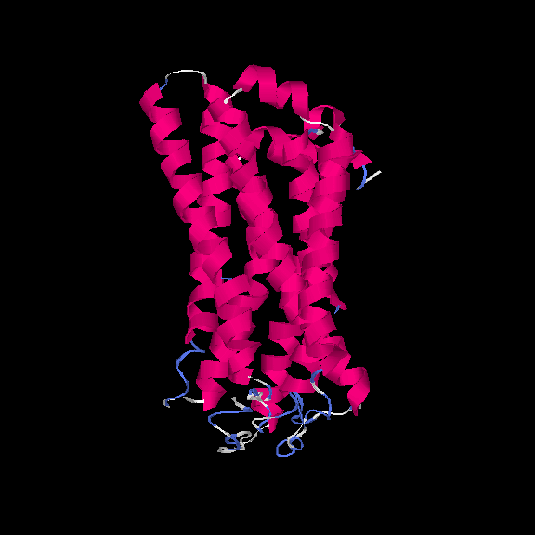 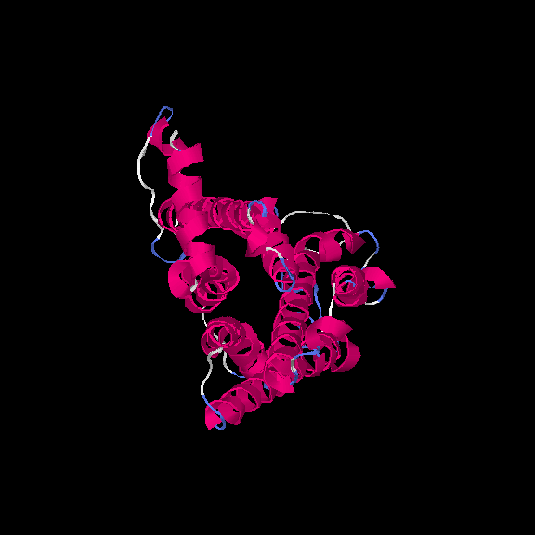 |
| HG0172 | O15354 | 613 | 0.35 | 0.76 ± 0.1 | 5.9 ± 3.7 |  |
| HG0173 | P43116 | 358 | -0.07 | 0.7 ± 0.12 | 6.7 ± 4 | 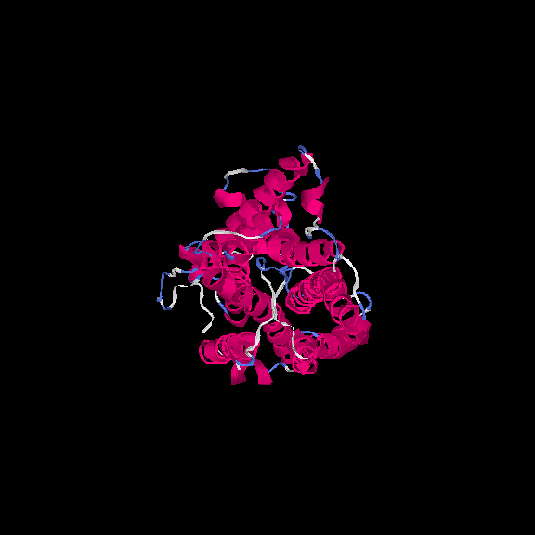 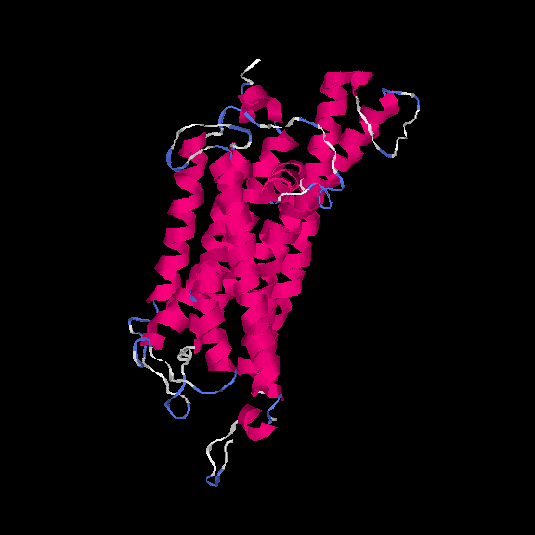 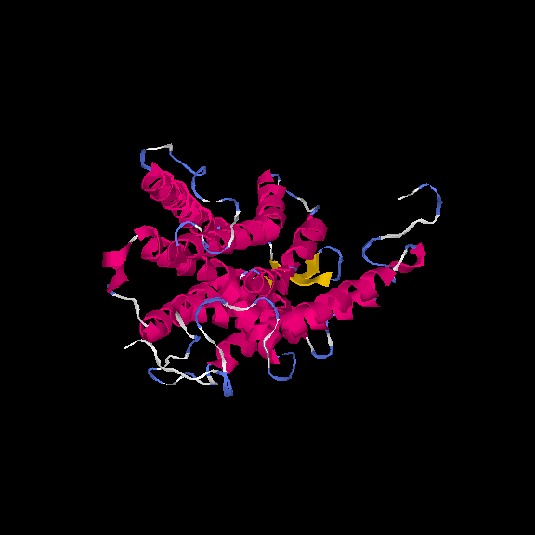 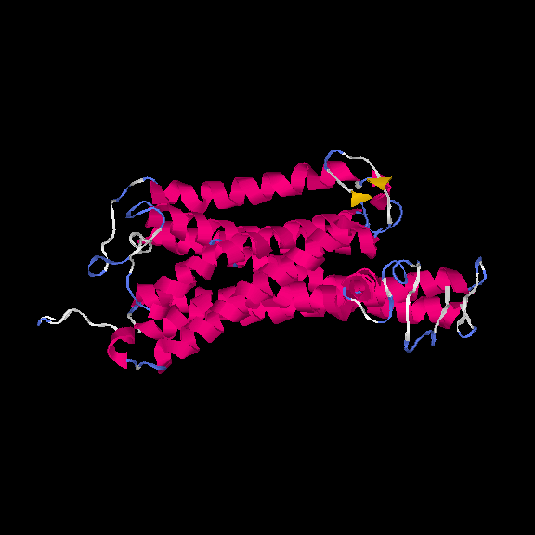 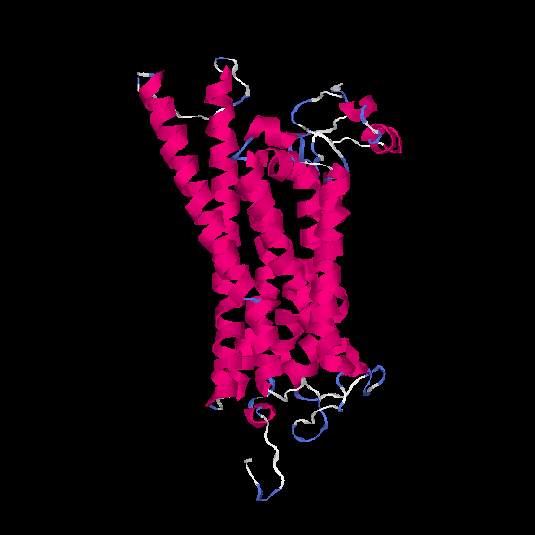 |
| HG0174 | Q6IFH4 | 312 | -0.21 | 0.69 ± 0.12 | 6.7 ± 4 | 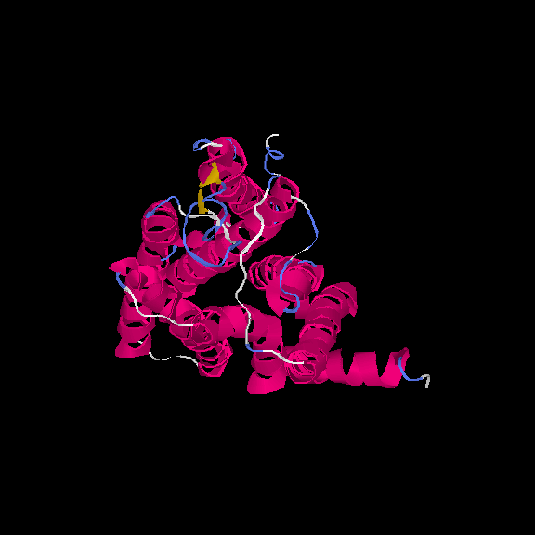 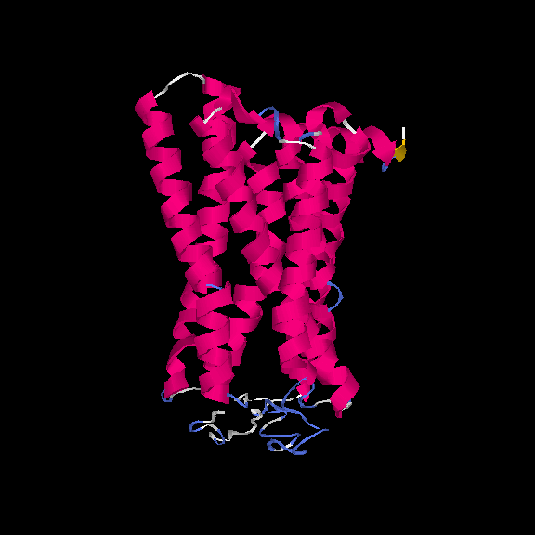 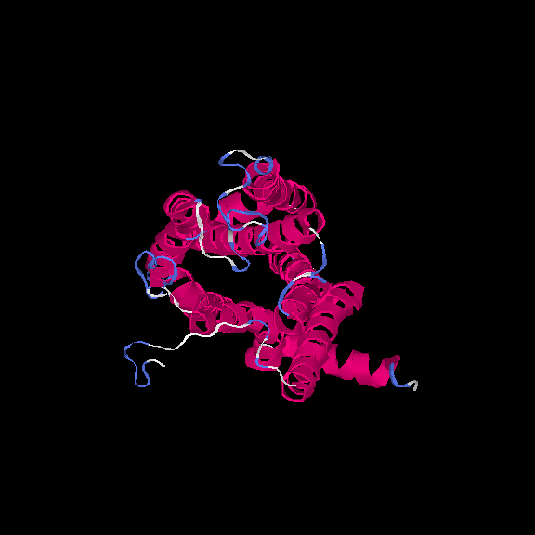 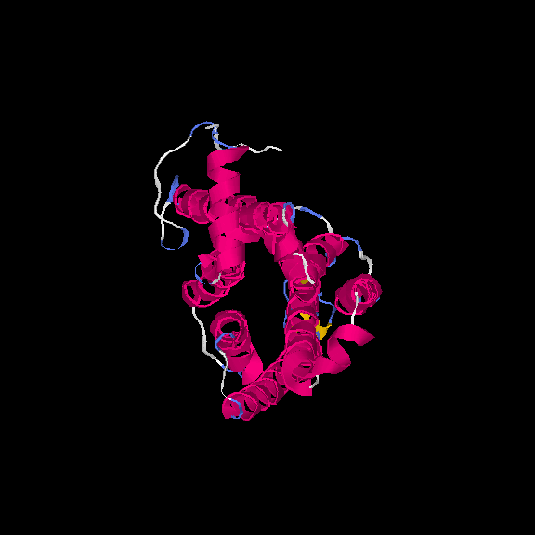 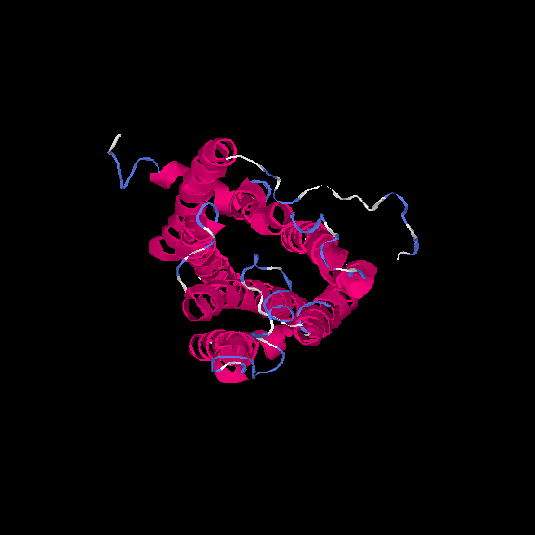 |
| HG0175 | Q96R09 | 309 | 0.12 | 0.73 ± 0.11 | 6 ± 3.7 | 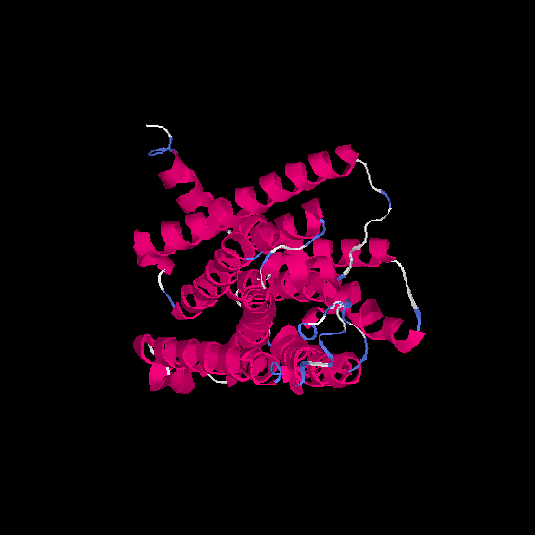 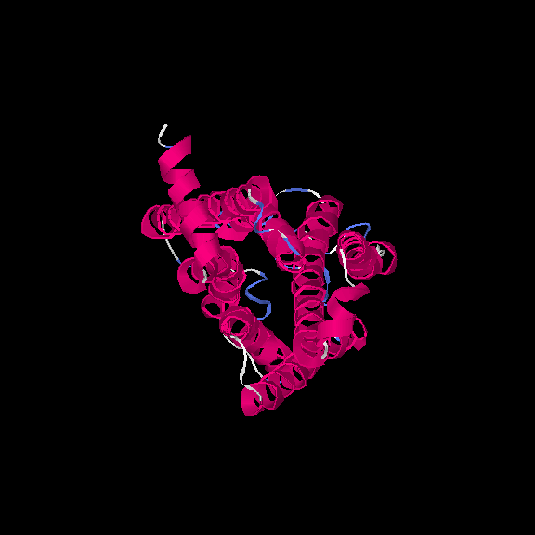   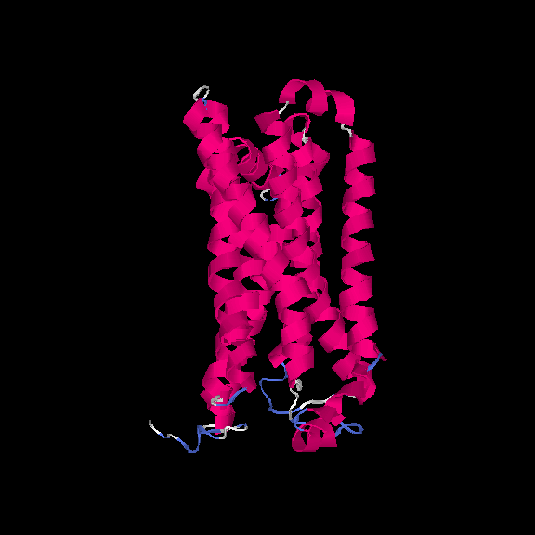 |
| HG0176 | P41586 | 468 | -0.87 | 0.6 ± 0.14 | 9.2 ± 4.6 | 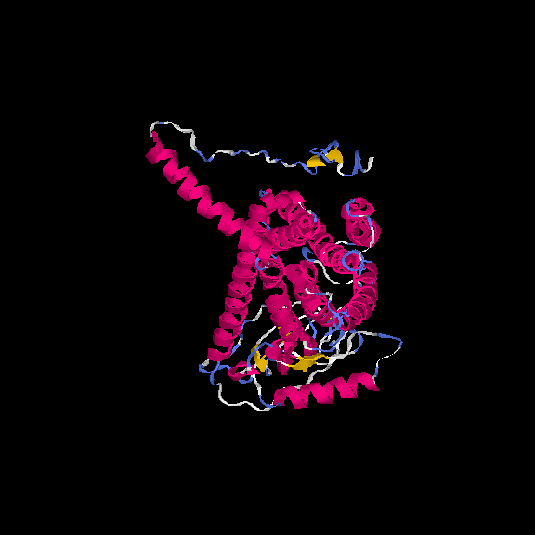 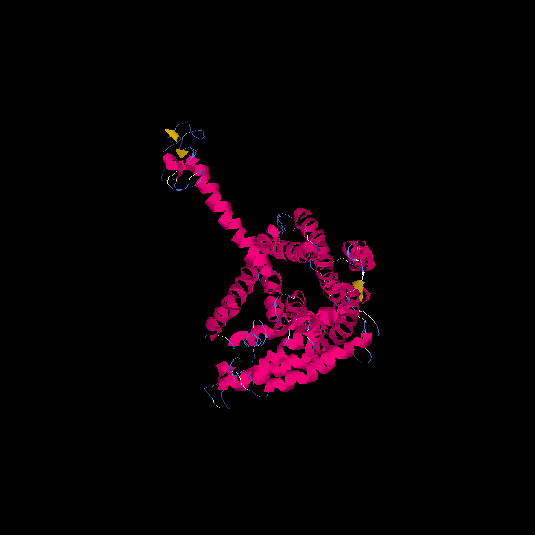   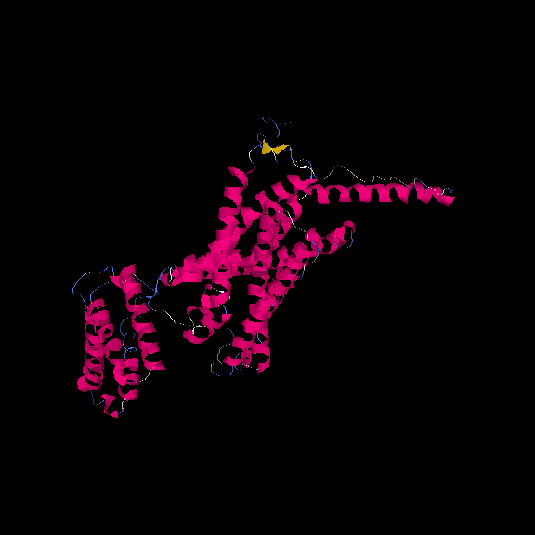 |
| HG0177 | A8KAG8 | 408 | -1.3 | 0.55 ± 0.15 | 9.8 ± 4.6 | 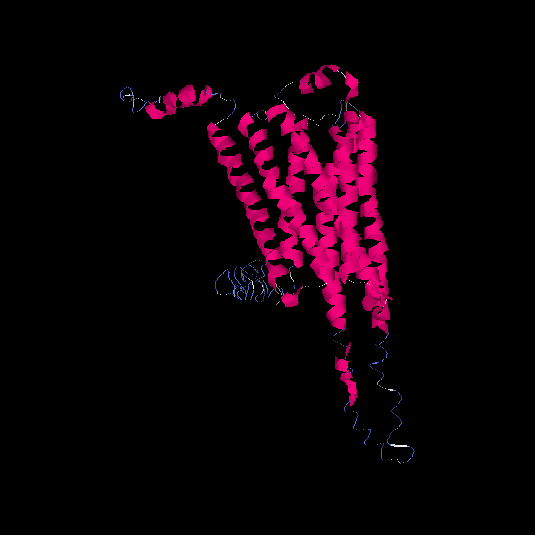 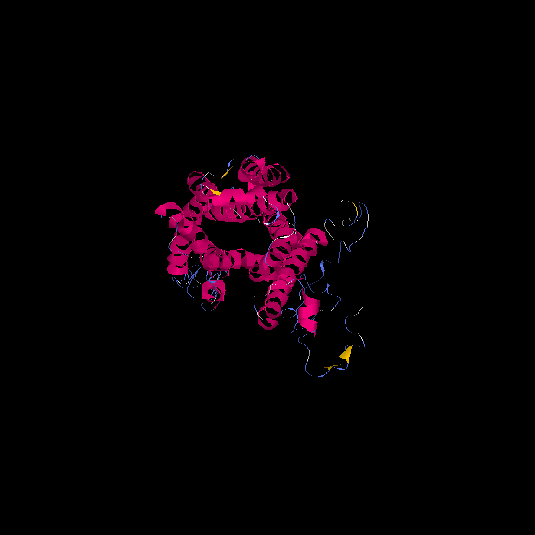 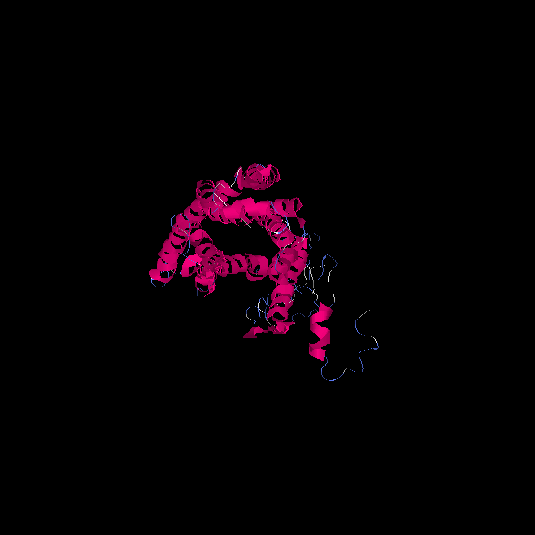  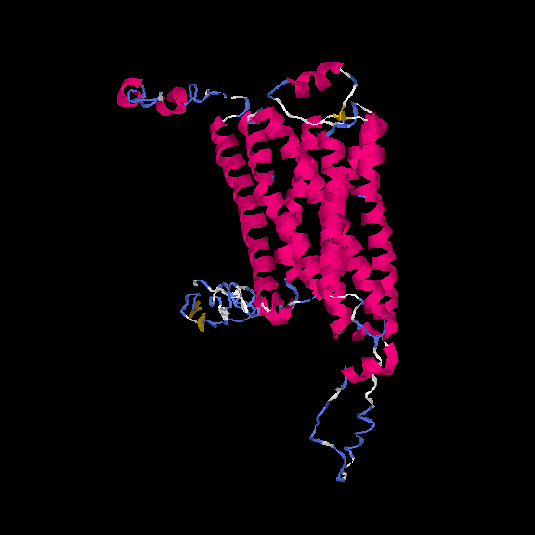 |
| HG0178 | Q8NGL3 | 314 | 0.1 | 0.73 ± 0.11 | 6.1 ± 3.8 | 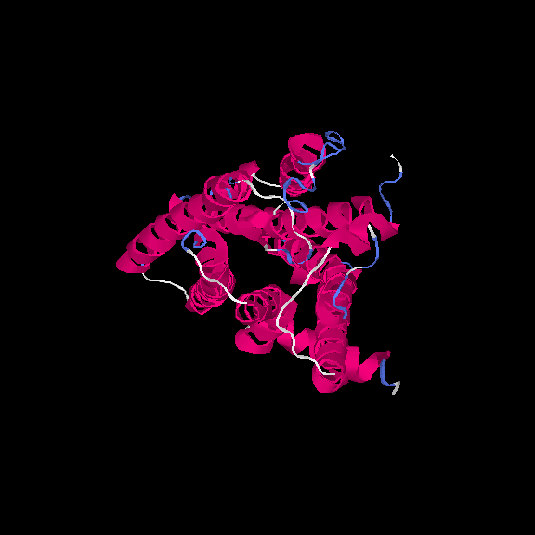 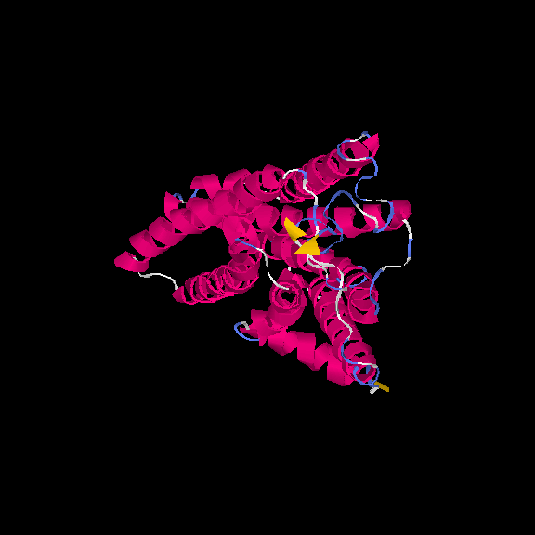 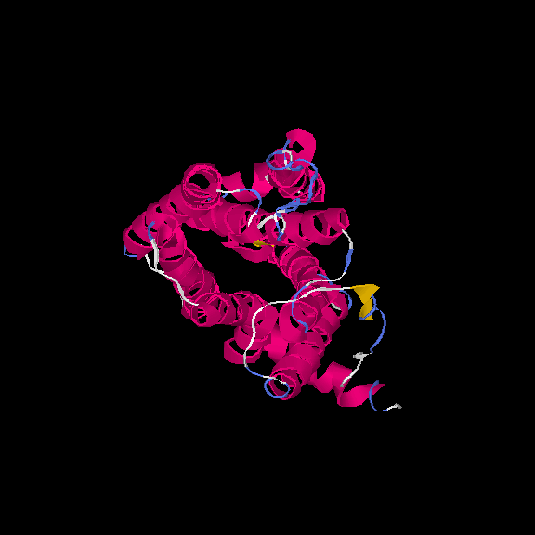 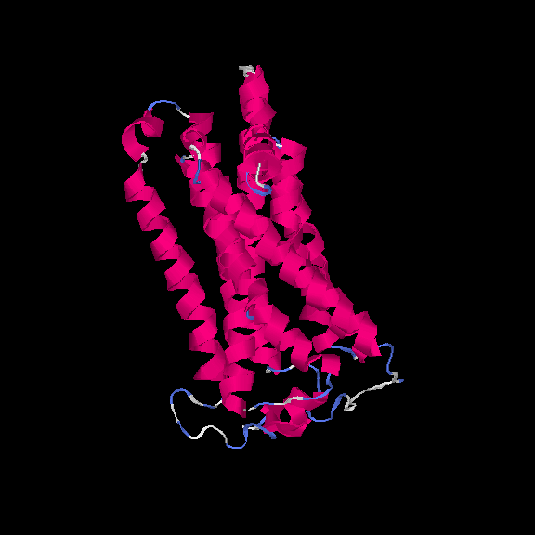 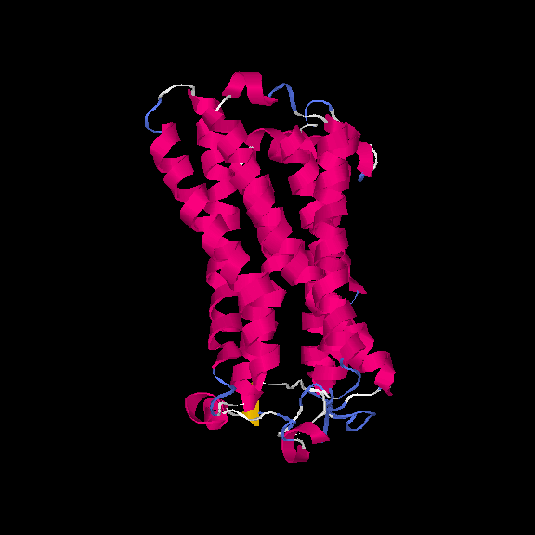 |
| HG0179 | Q8NGL9 | 310 | -0.05 | 0.71 ± 0.12 | 6.4 ± 3.9 | 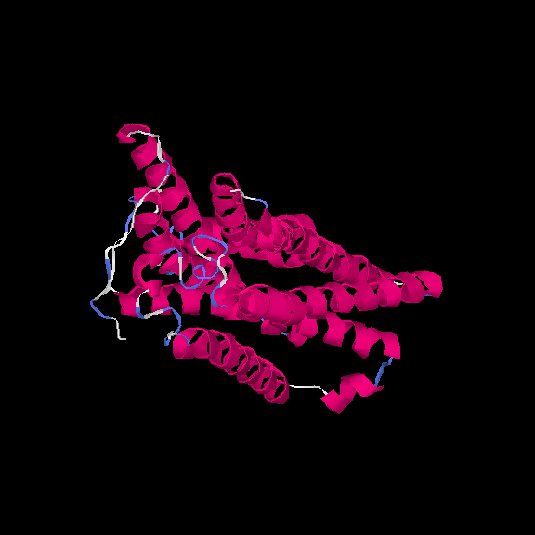 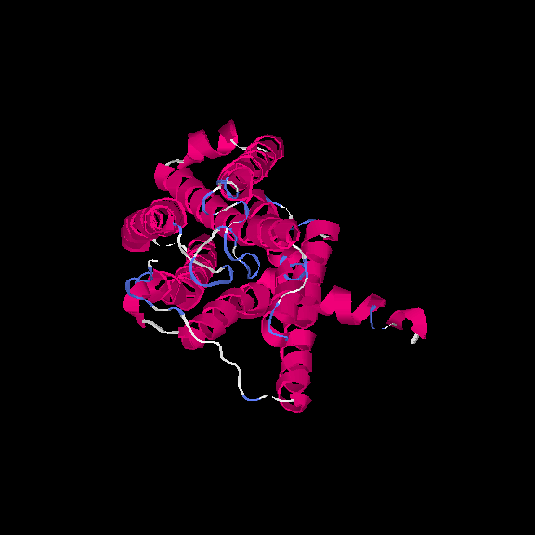 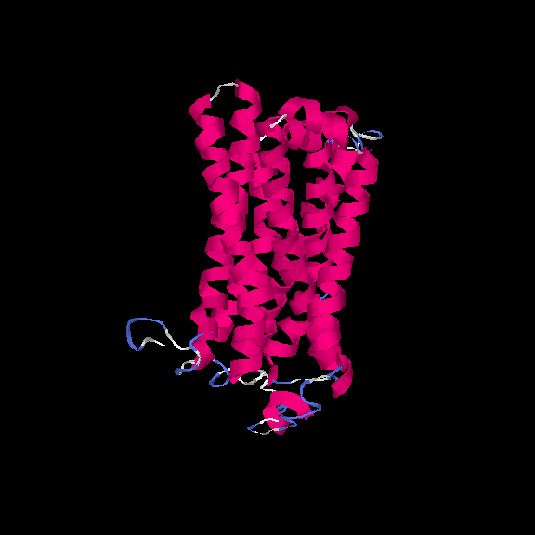 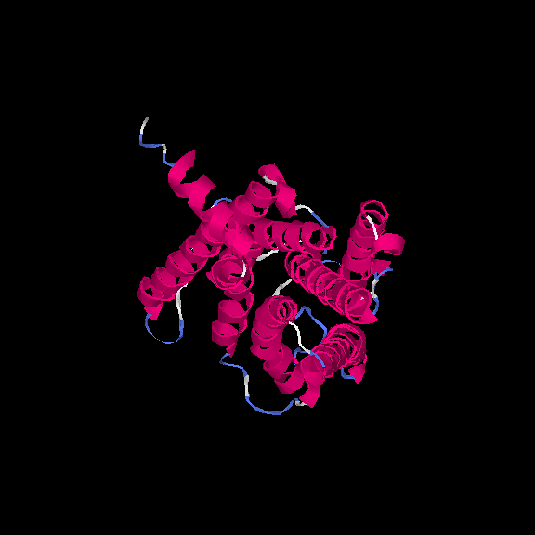 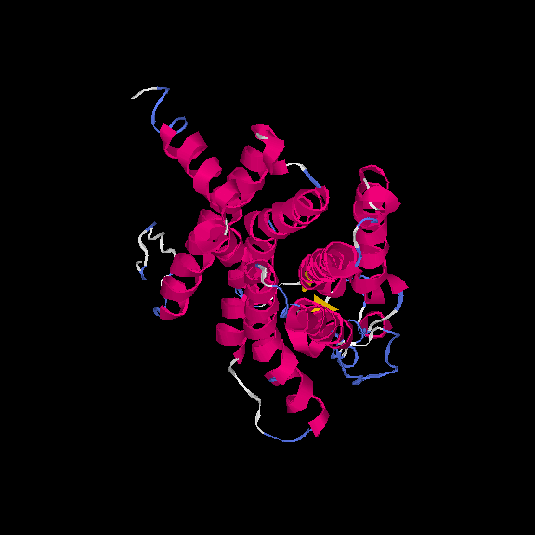 |
| HG0180 | Q9Y5P0 | 310 | -0.01 | 0.71 ± 0.11 | 6.3 ± 3.8 | 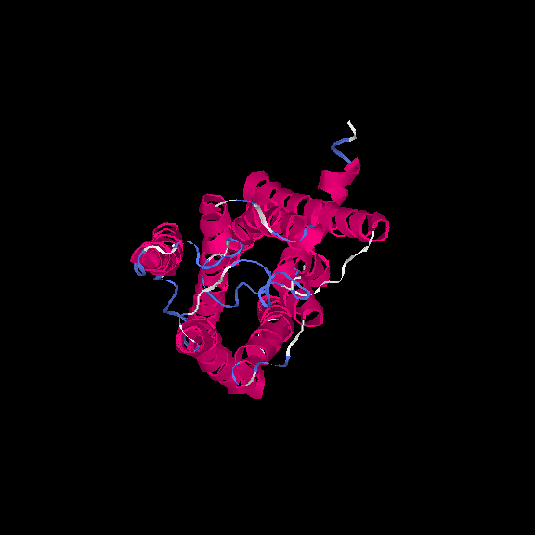 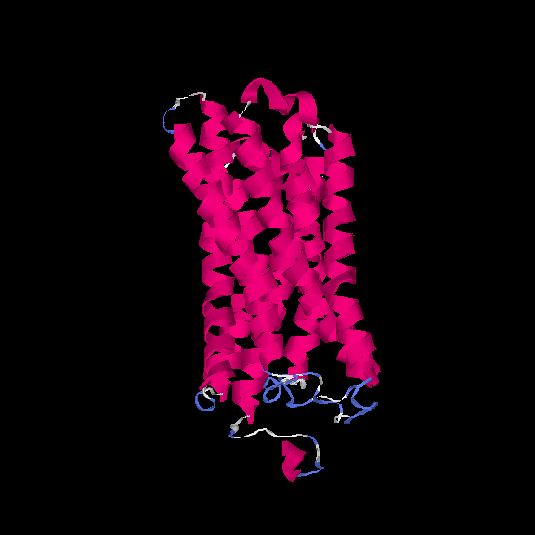 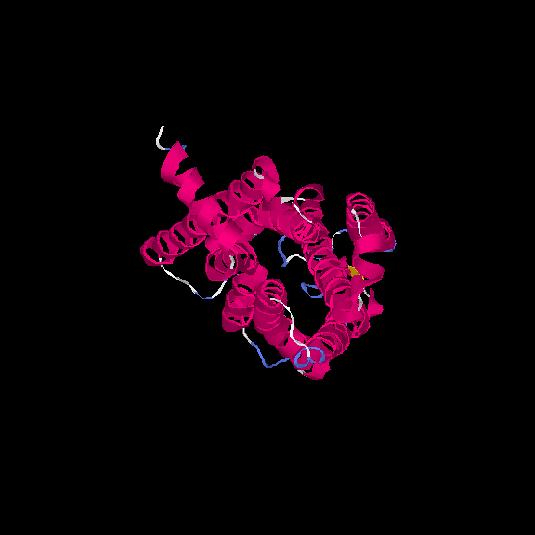 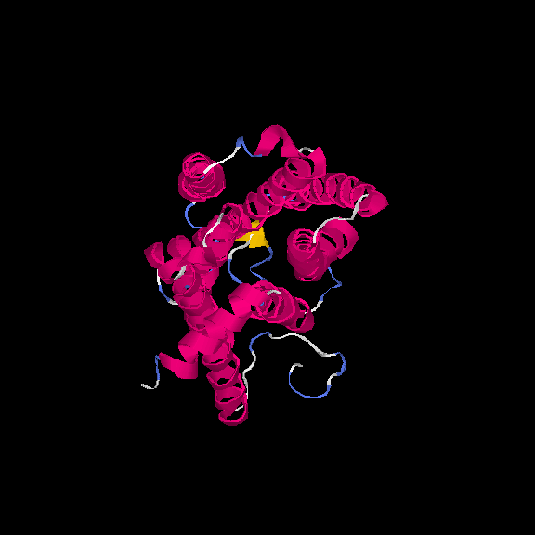  |
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218