


| HG ID | UniProt ID | Length | C-score | Estimated TM-score |
Estimated RMSD |
Top 5 models |
| HG0120 | Q96RI0 | 385 | 0.38 | 0.76 ± 0.1 | 5.9 ± 3.7 | 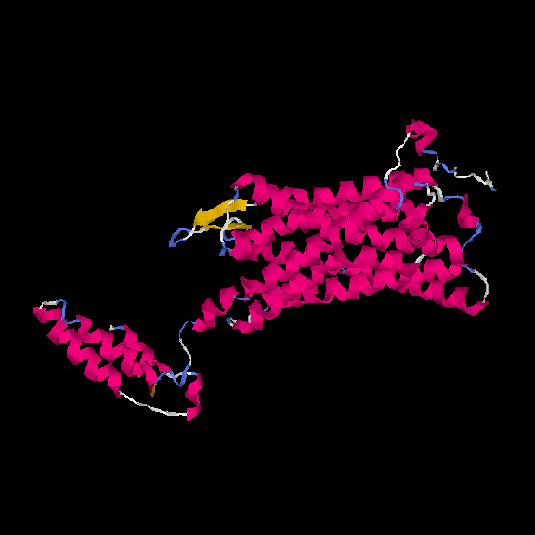 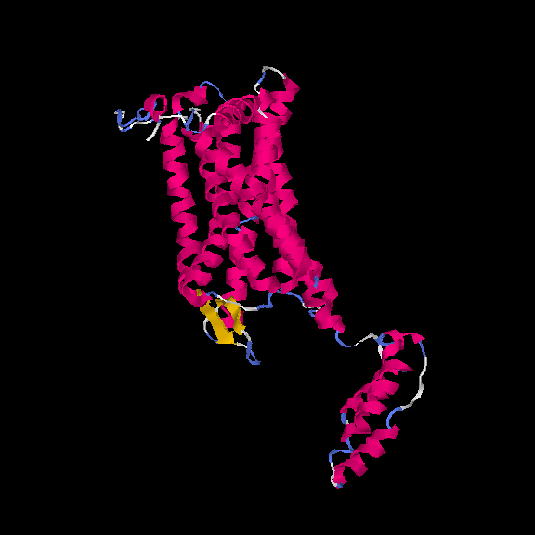 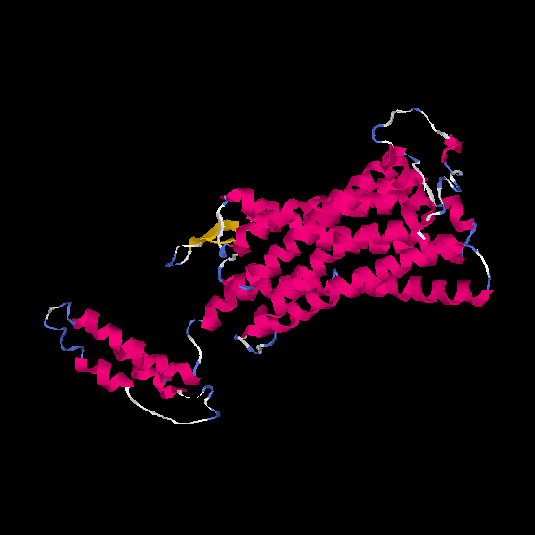 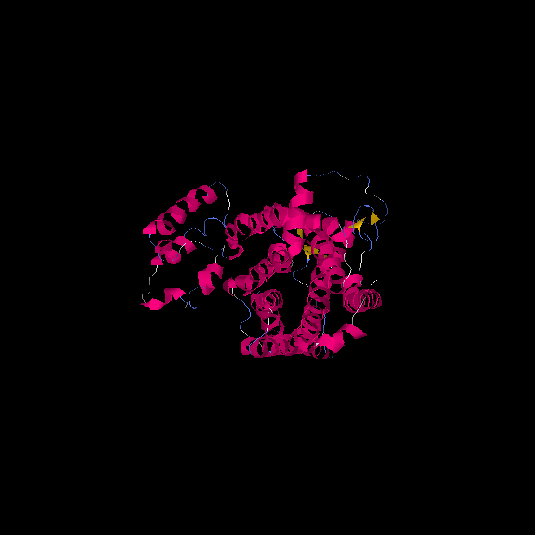 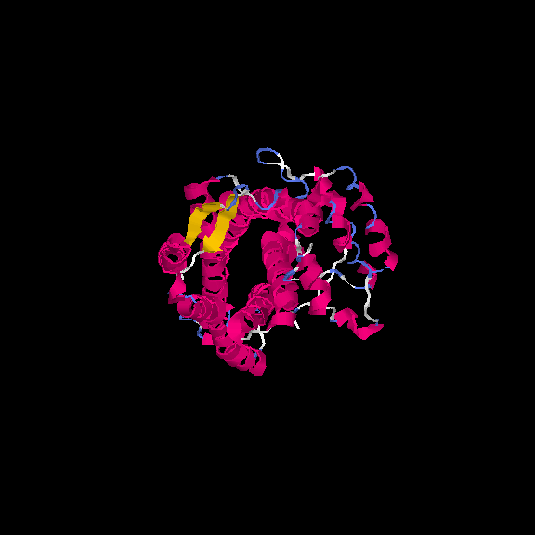 |
| HG0121 | Q8NH61 | 342 | -1.07 | 0.58 ± 0.14 | 8.9 ± 4.6 | 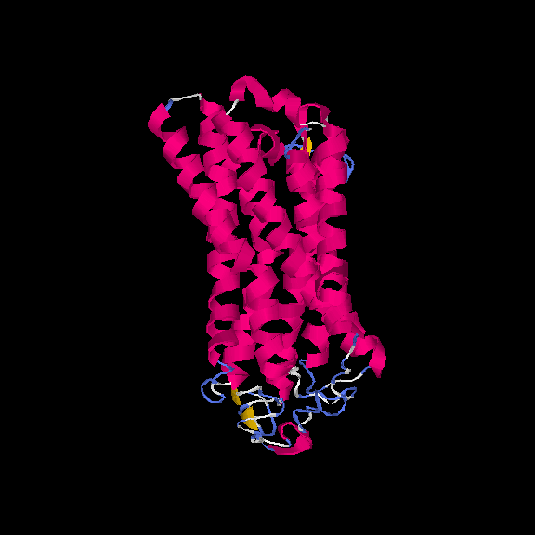 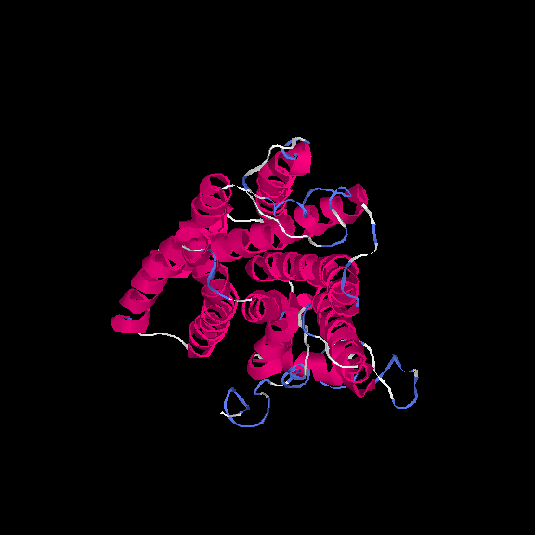 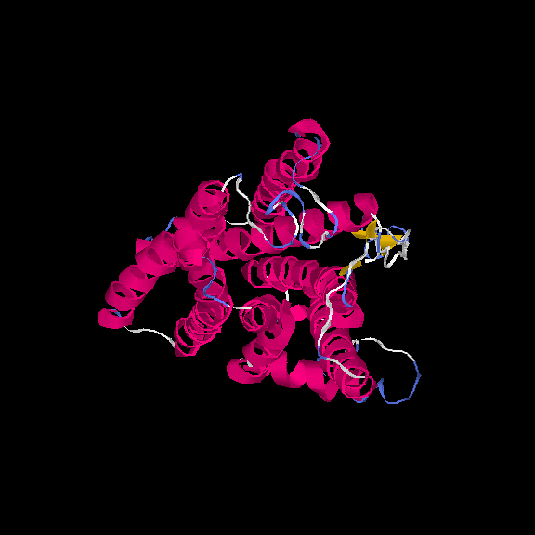 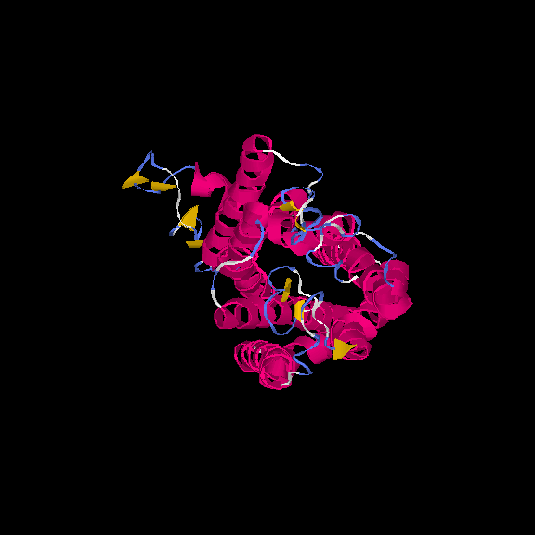 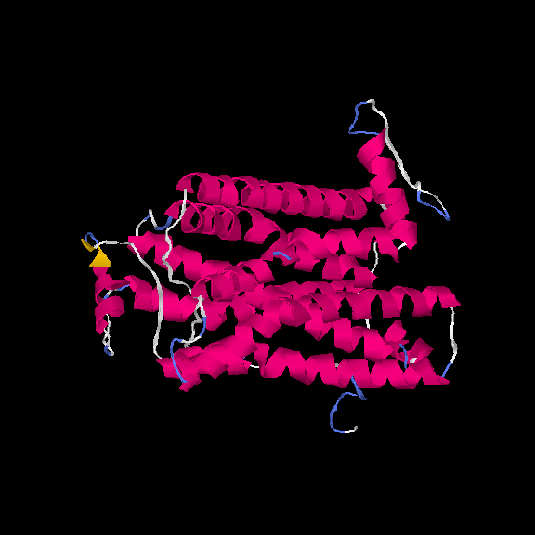 |
| HG0122 | Q8WZ94 | 311 | 0.09 | 0.72 ± 0.11 | 6.1 ± 3.8 | 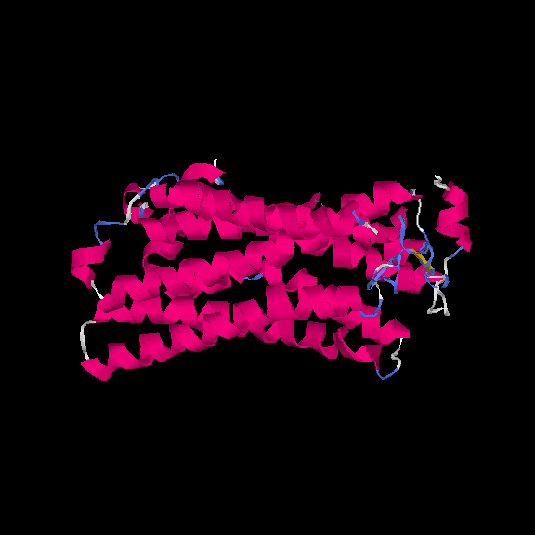 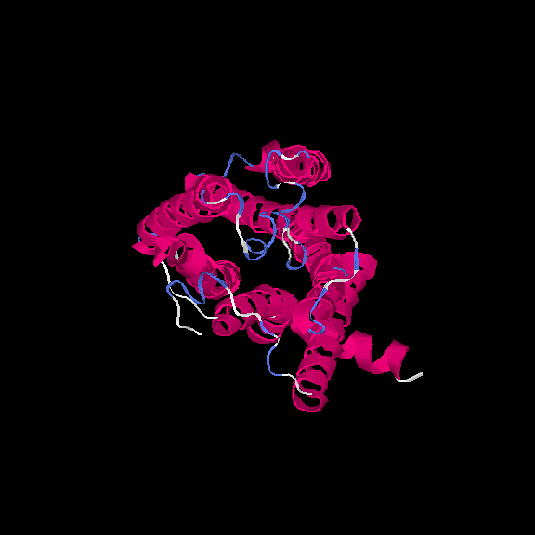 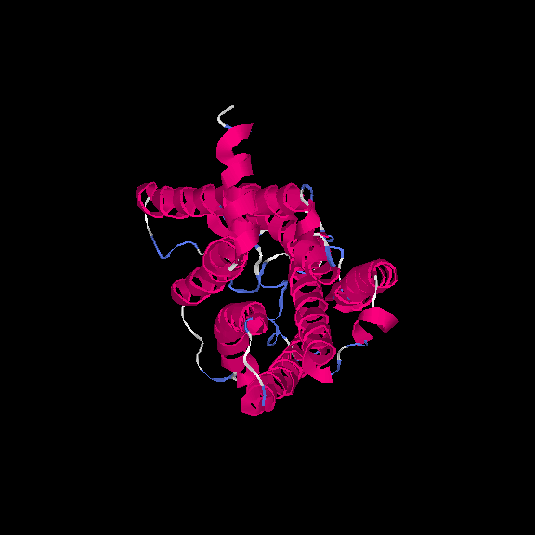 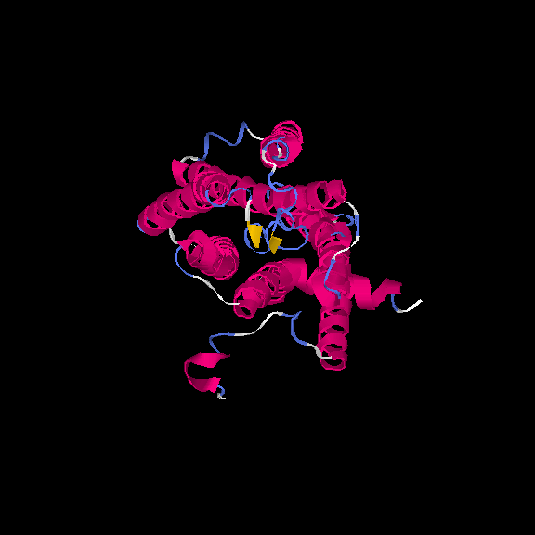 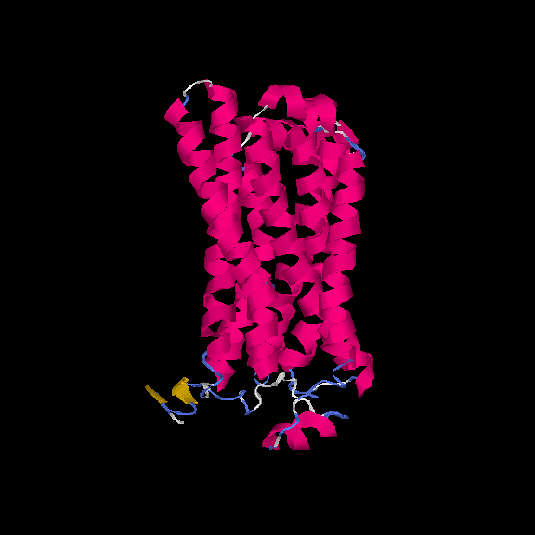 |
| HG0123 | A8K2H7 | 482 | -0.48 | 0.65 ± 0.13 | 7.5 ± 4.3 | 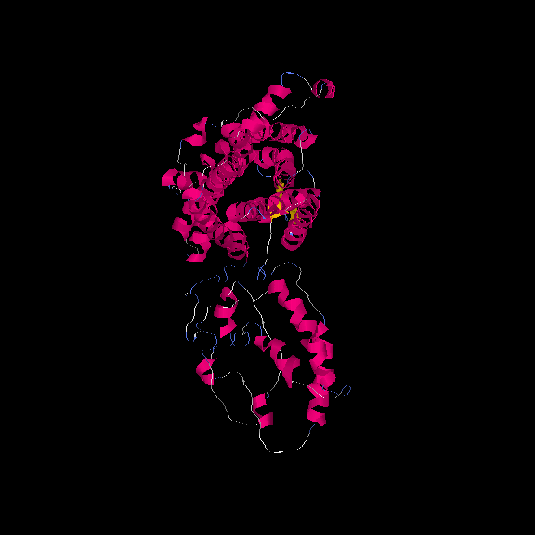 |
| HG0124 | Q96P68 | 337 | 0.37 | 0.76 ± 0.1 | 5.7 ± 3.6 | 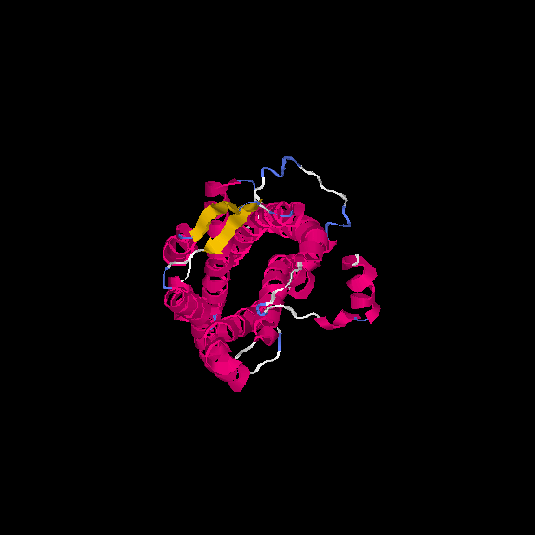  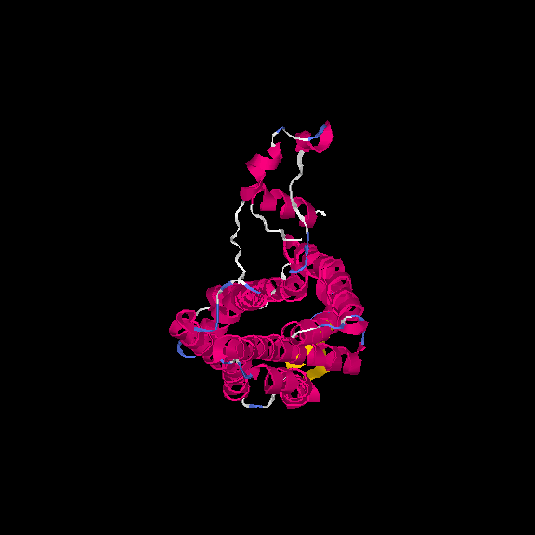 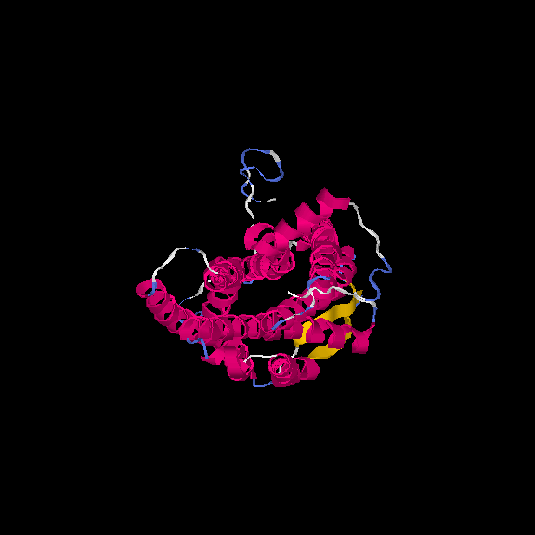 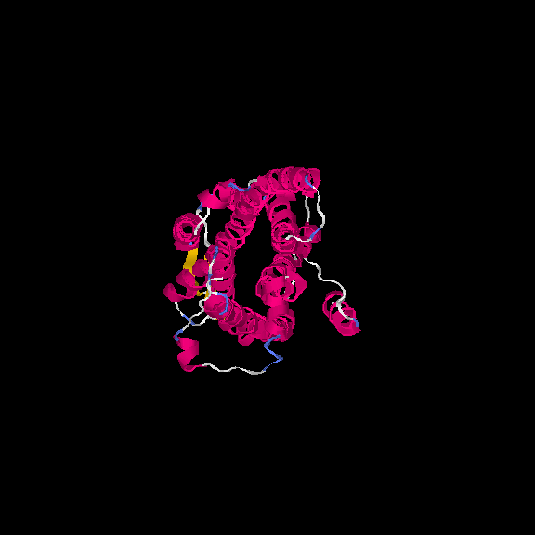 |
| HG0125 | Q5EKM8 | 352 | -0.94 | 0.6 ± 0.14 | 8.7 ± 4.5 | 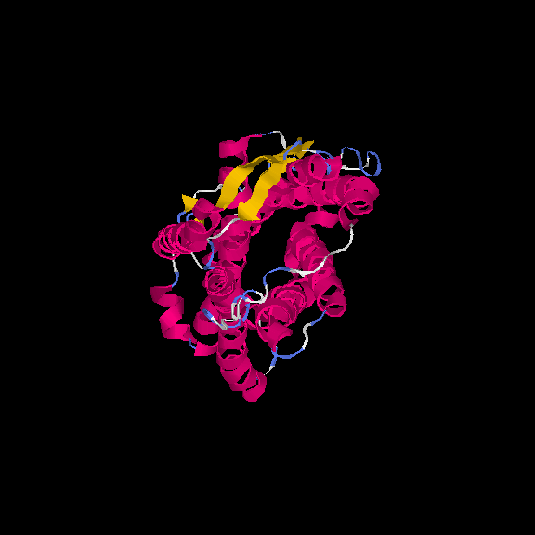 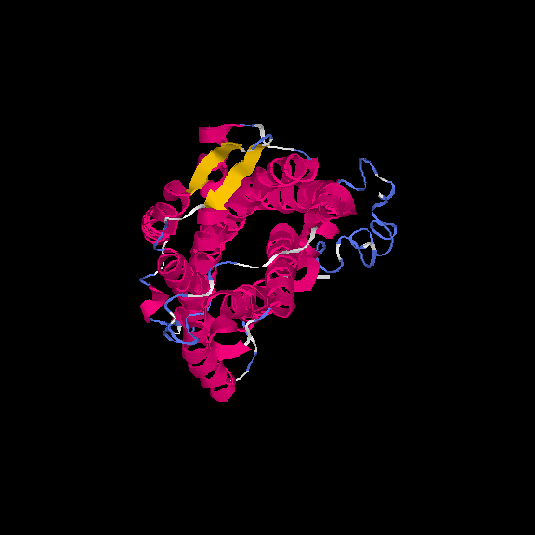 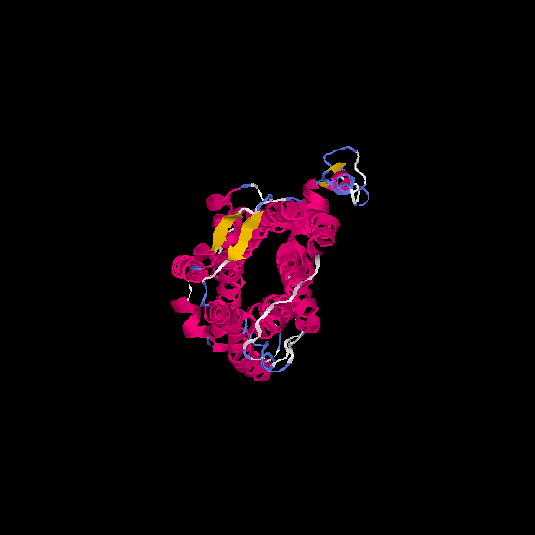 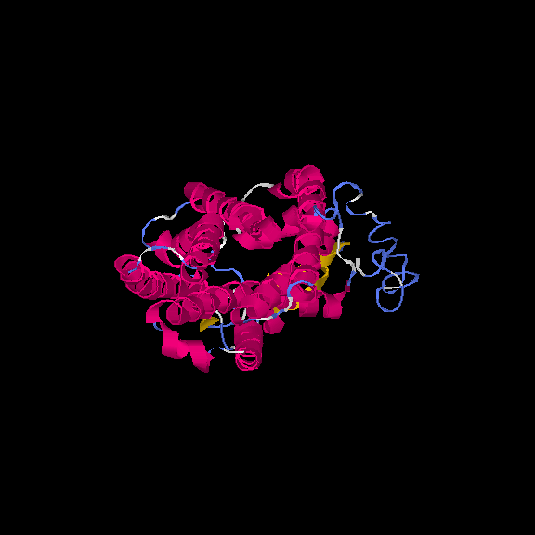 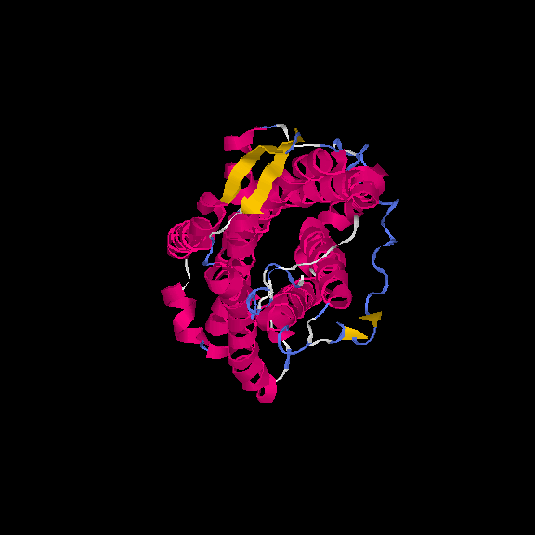 |
| HG0126 | P32245 | 332 | -0.1 | 0.7 ± 0.12 | 6.6 ± 4 | 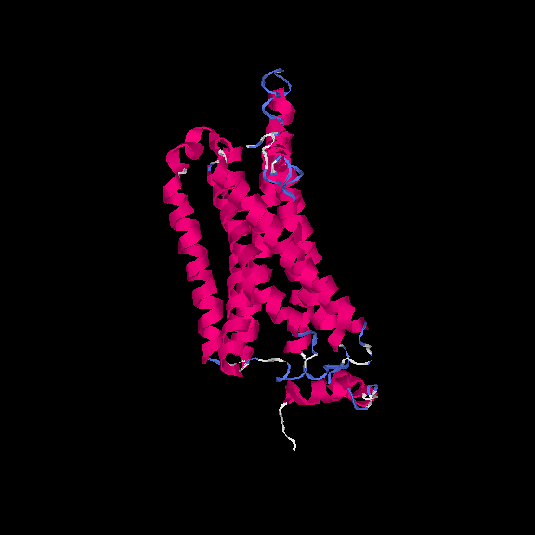 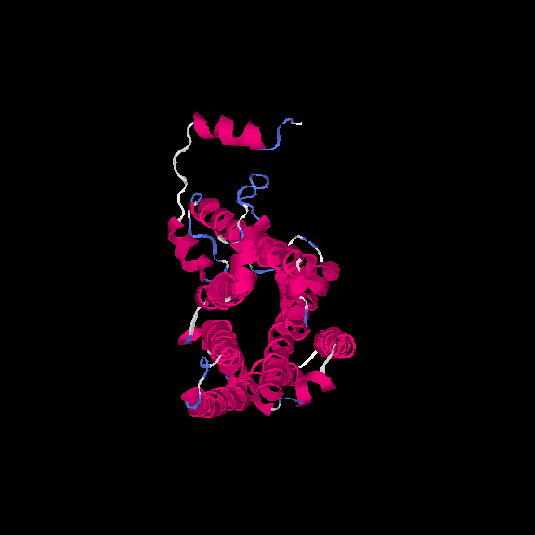 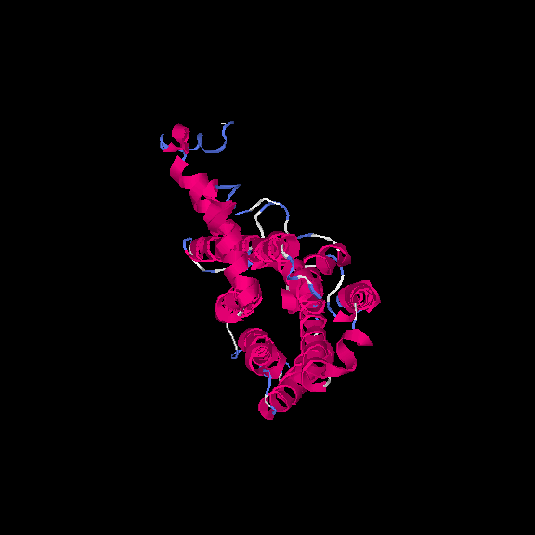 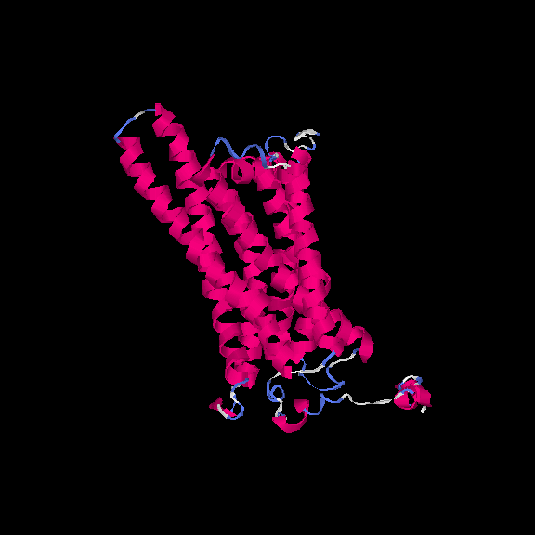 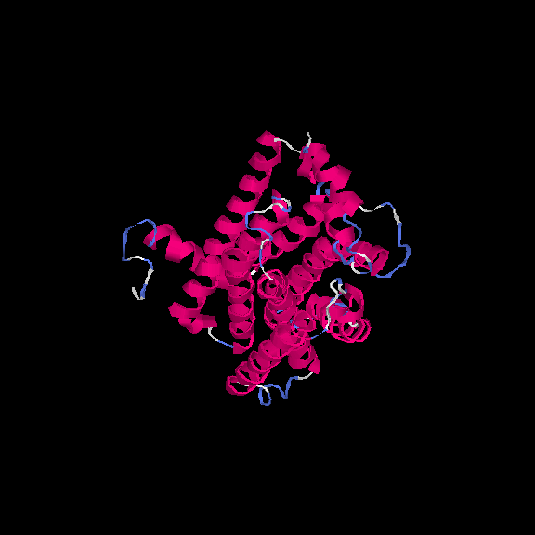 |
| HG0127 | Q4VBL0 | 390 | -0.6 | 0.64 ± 0.13 | 8.1 ± 4.4 | 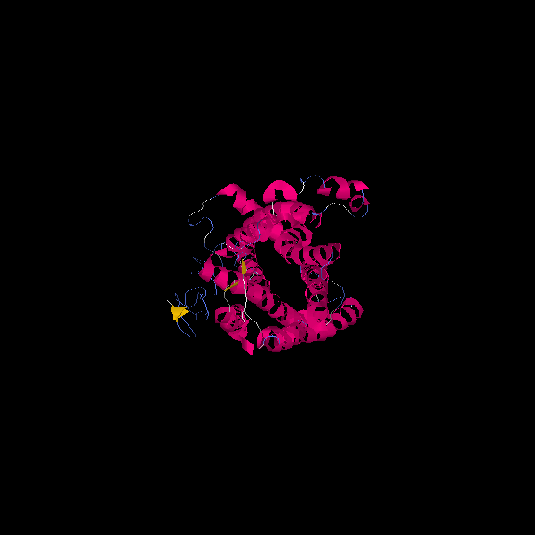 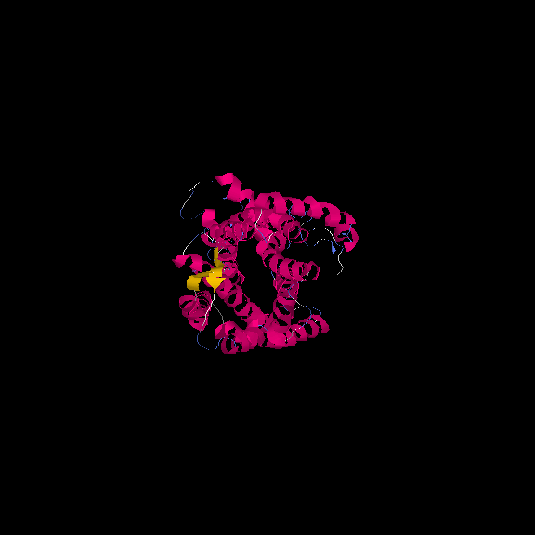 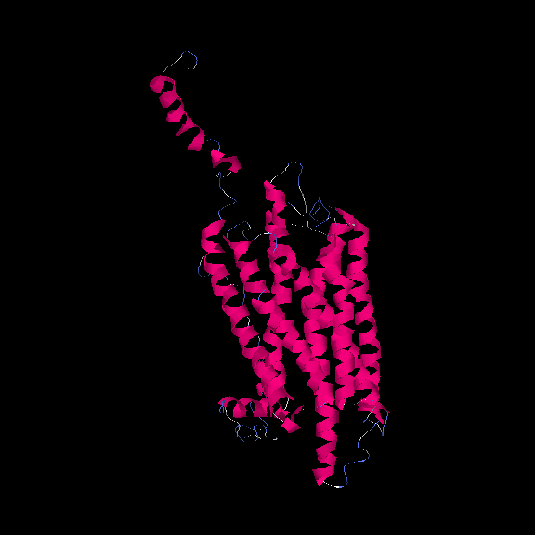  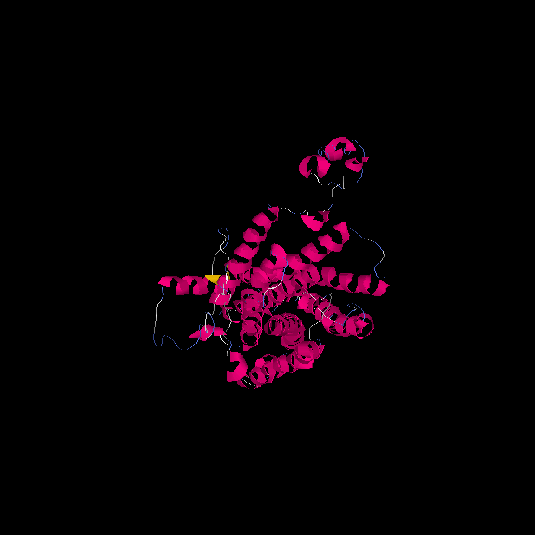 |
| HG0128 | Q8NGE0 | 317 | -0.31 | 0.67 ± 0.12 | 7 ± 4.1 | 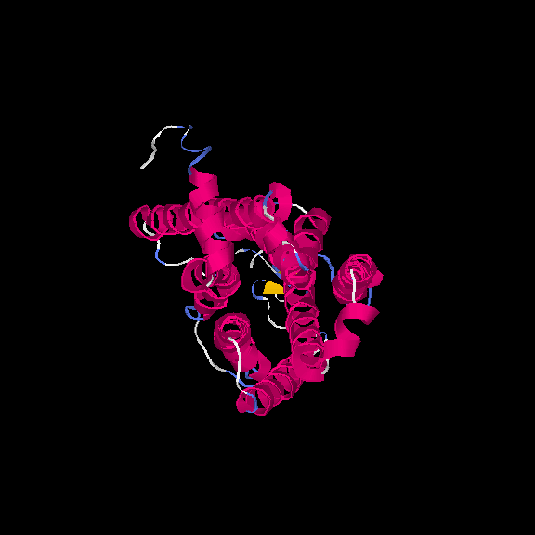 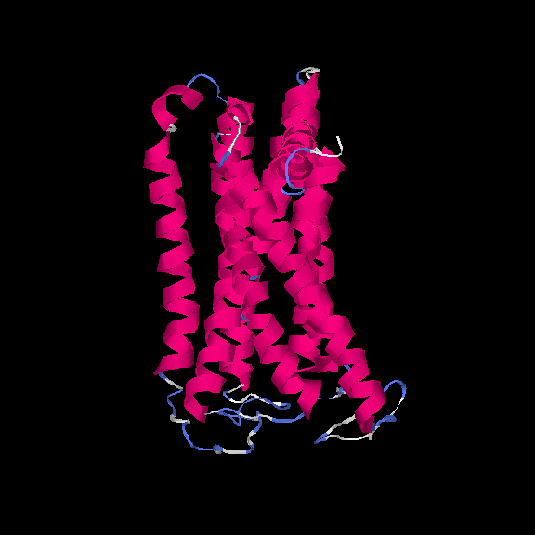 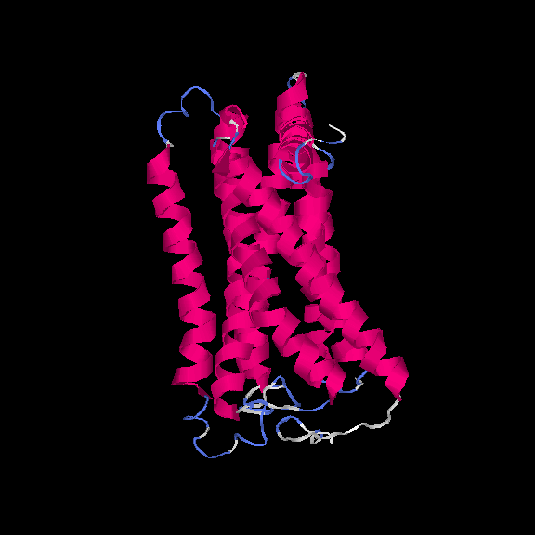 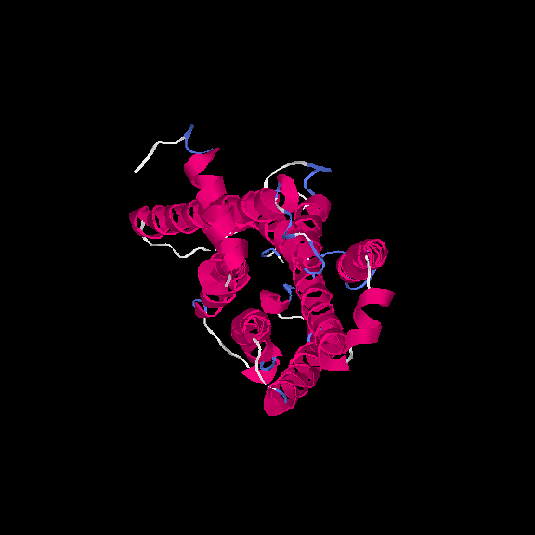 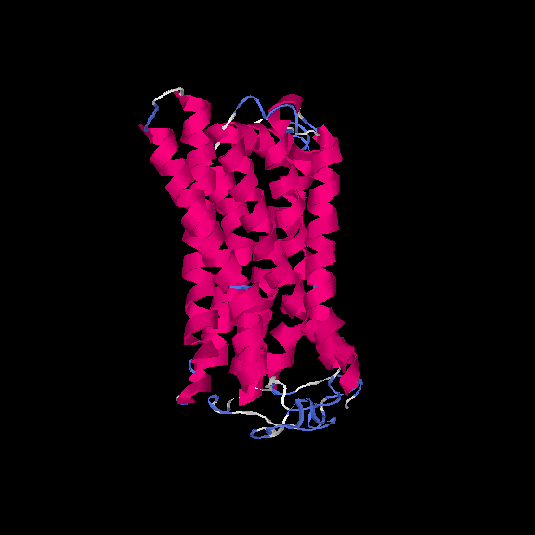 |
| HG0129 | Q86YG3 | 468 | 0.36 | 0.76 ± 0.1 | 4.2 ± 2.8 |  |
| HG0130 | Q8NGU4 | 316 | -0.13 | 0.7 ± 0.12 | 6.6 ± 4 | 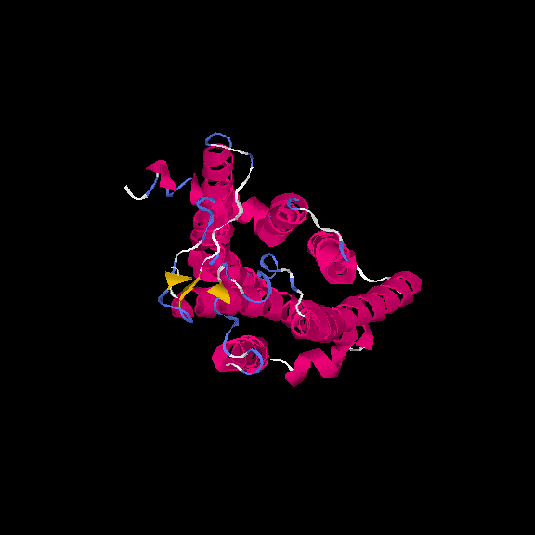 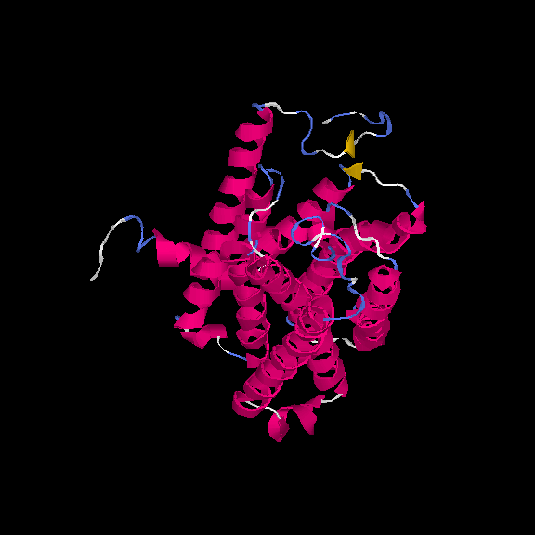 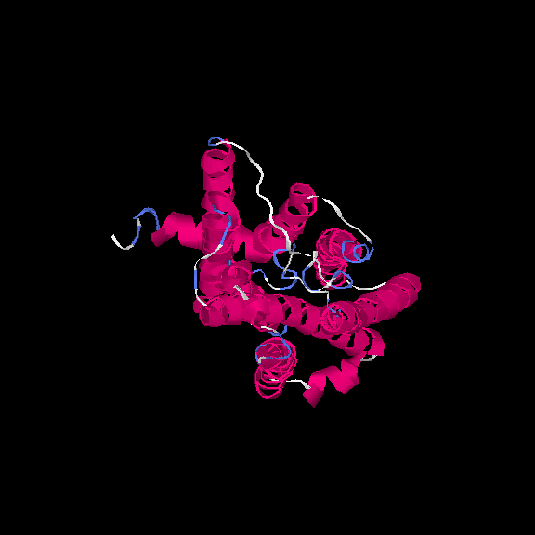 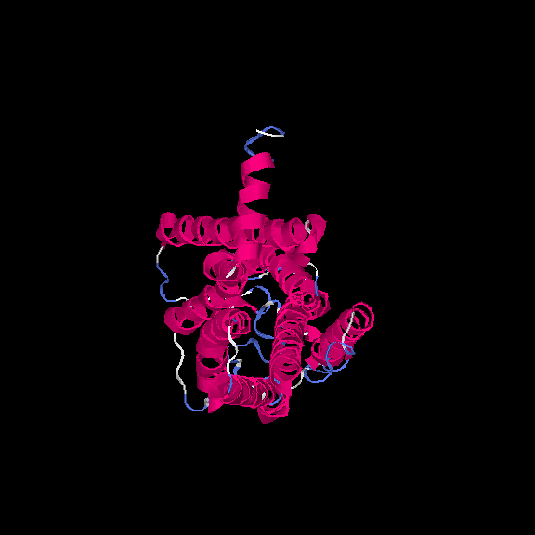 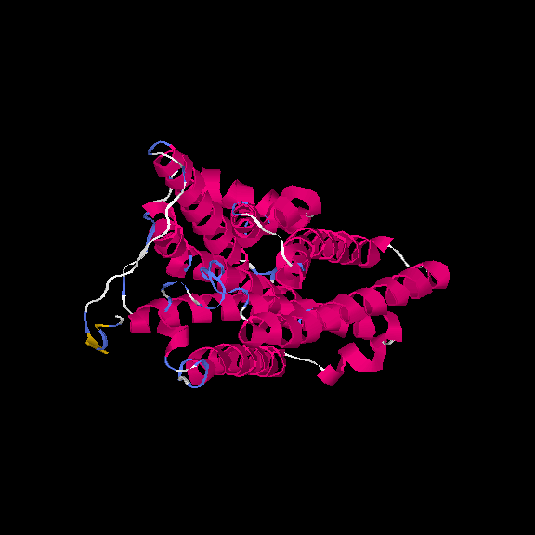 |
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218