


| HG ID | UniProt ID | Length | C-score | Estimated TM-score |
Estimated RMSD |
Top 5 models |
| HG0020 | Q9Y5X5 | 522 | -2.2 | 0.45 ± 0.15 | 9.99 ± 4.2 | 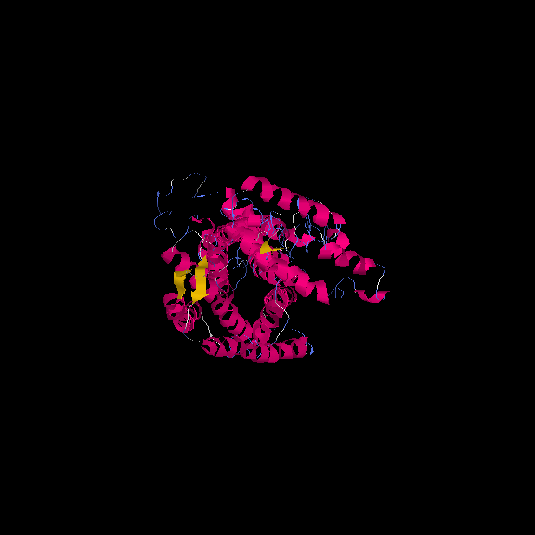 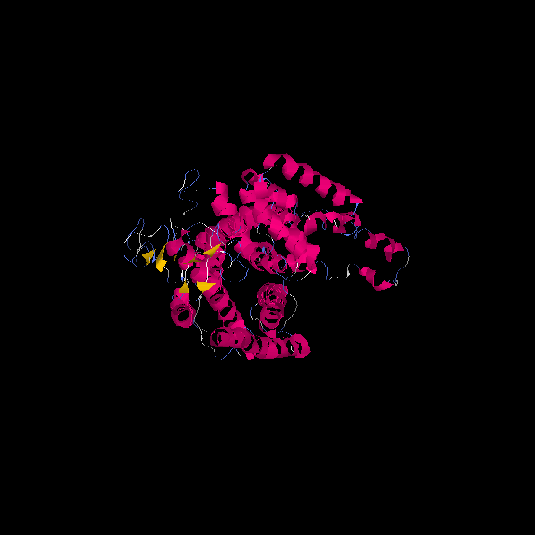 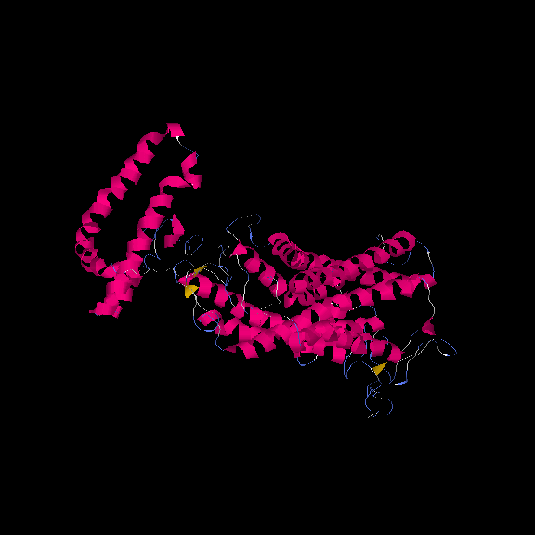 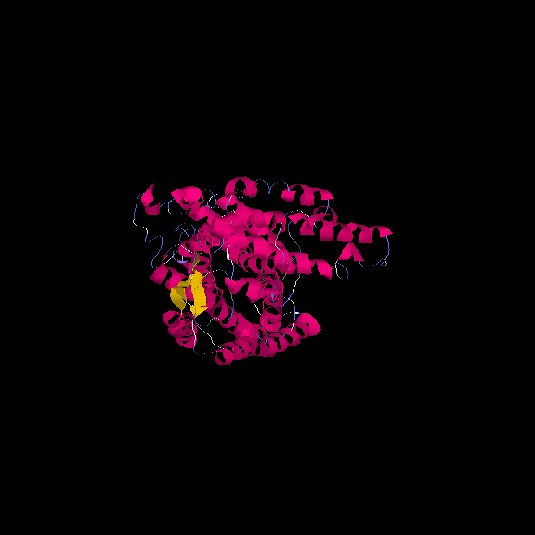  |
| HG0021 | Q6IFR1 | 324 | -0.72 | 0.62 ± 0.14 | 7.9 ± 4.4 |    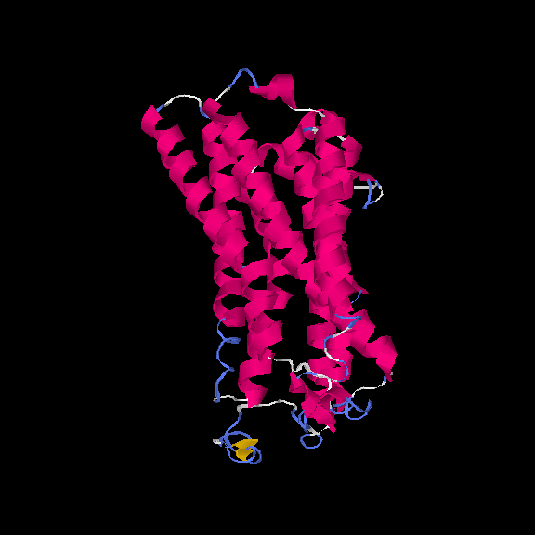 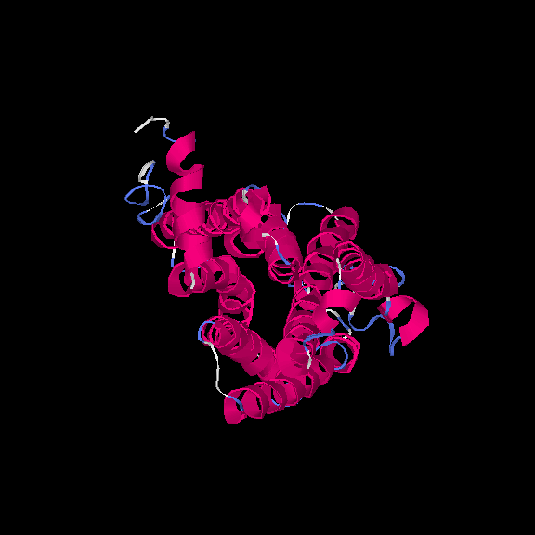 |
| HG0022 | Q8NGK1 | 321 | -0.42 | 0.66 ± 0.13 | 7.3 ± 4.2 |  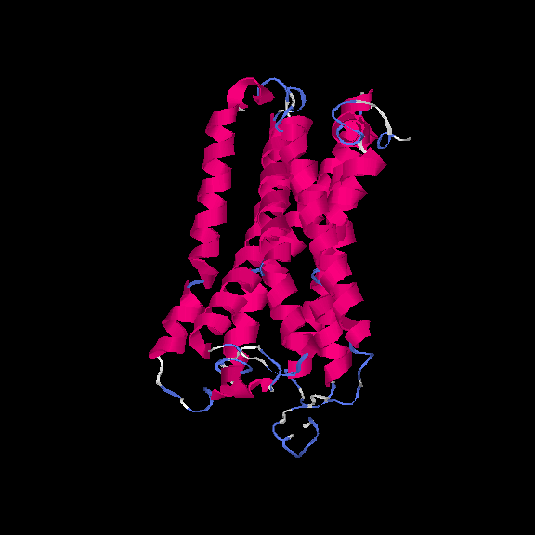 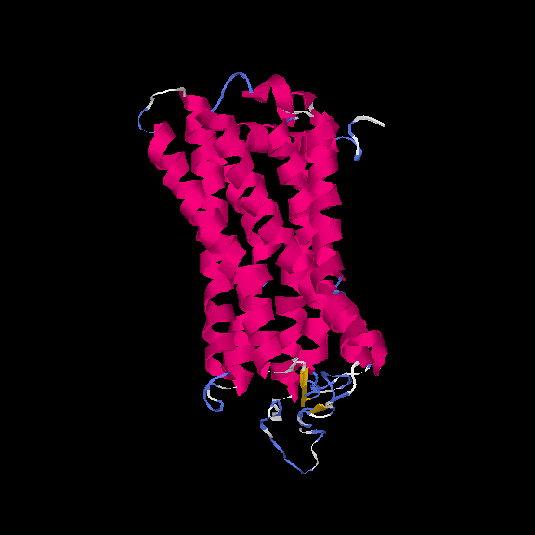 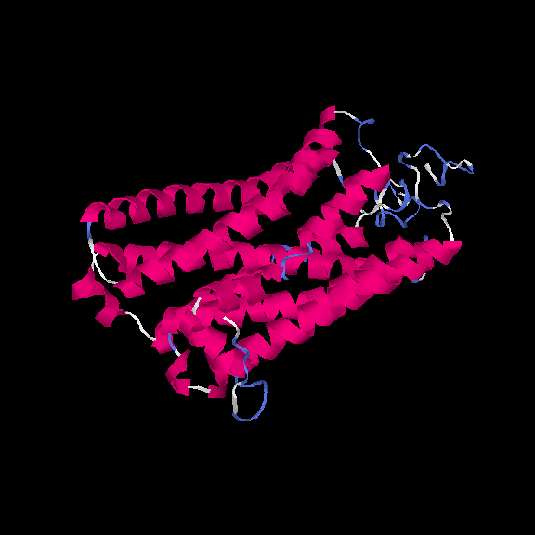 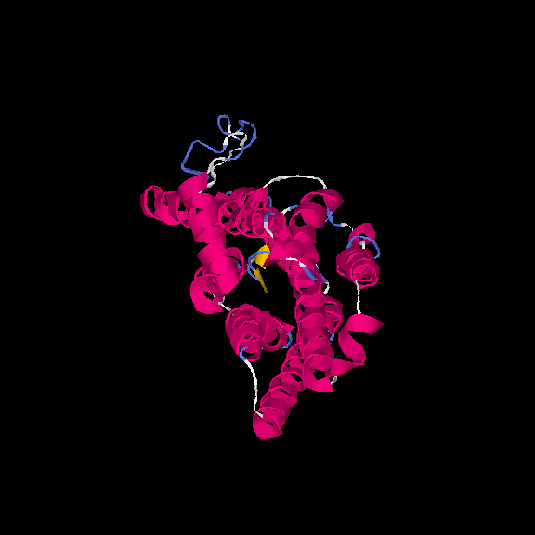 |
| HG0023 | Q8NGJ0 | 315 | -0.11 | 0.7 ± 0.12 | 6.5 ± 3.9 | 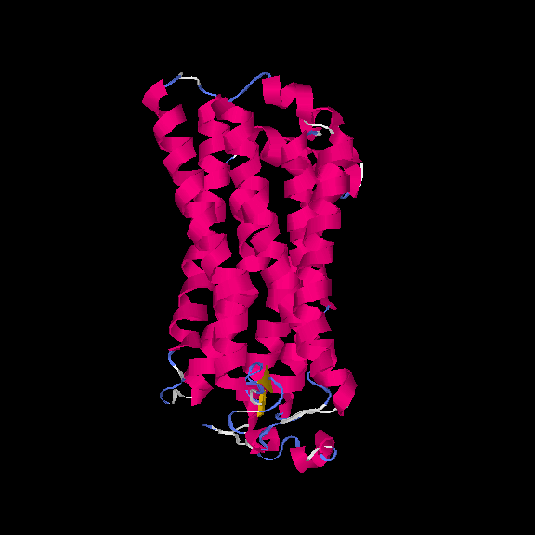 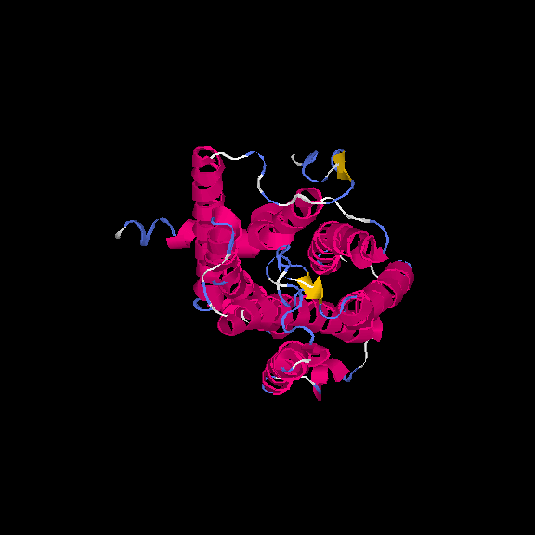    |
| HG0024 | P47884 | 311 | 0 | 0.71 ± 0.11 | 6.3 ± 3.8 |  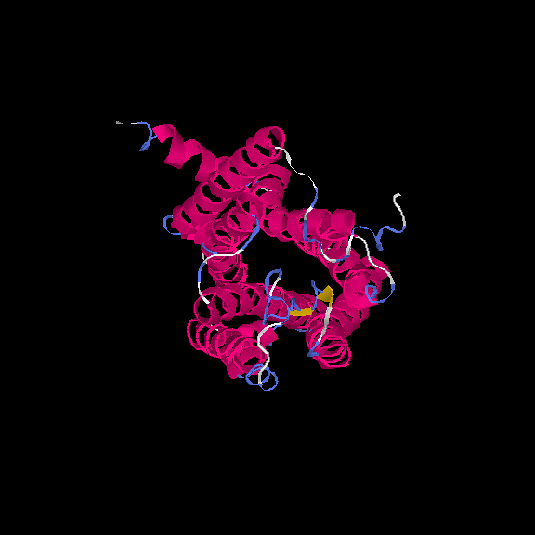 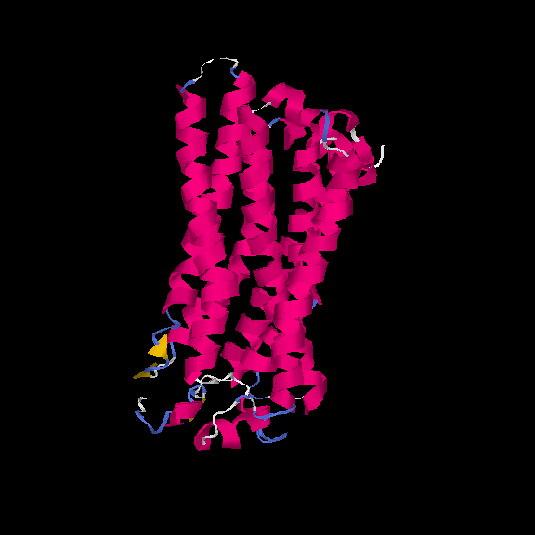 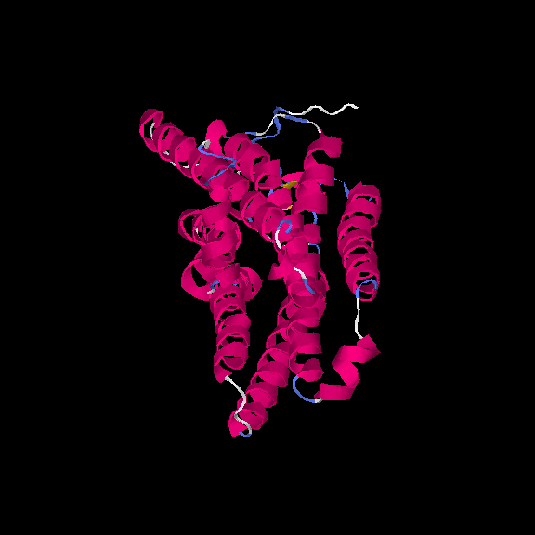  |
| HG0025 | Q96RI9 | 348 | -0.2 | 0.69 ± 0.12 | 6.9 ± 4.1 | 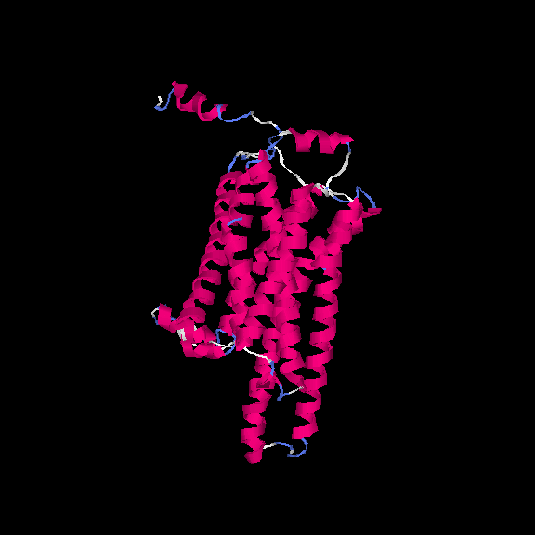 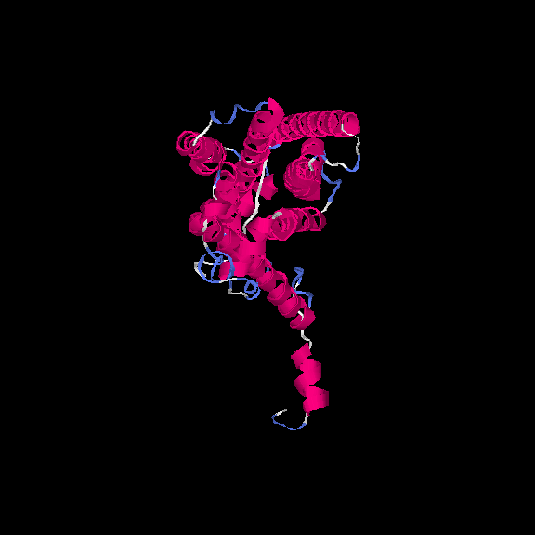 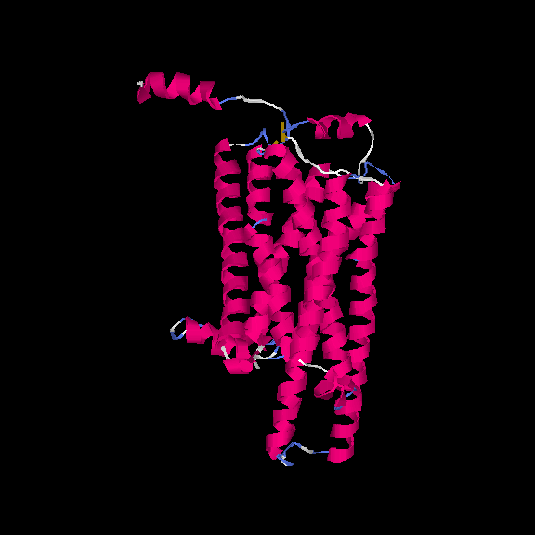 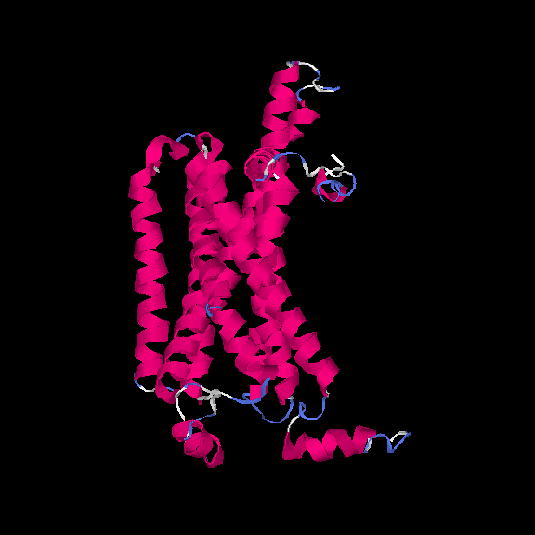 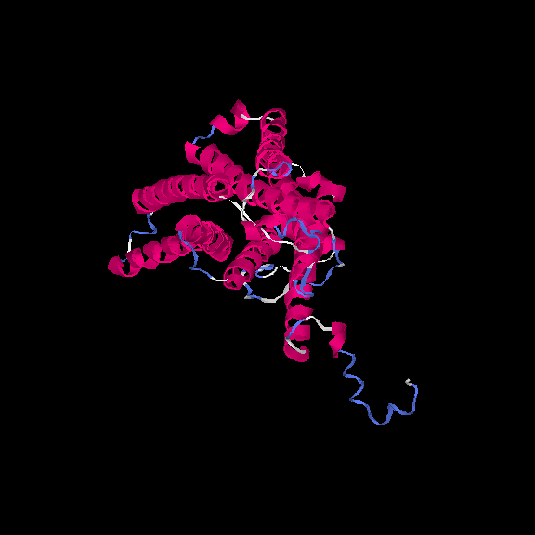 |
| HG0026 | P29371 | 465 | -2.5 | 0.42 ± 0.14 | 9.99 ± 4.1 |   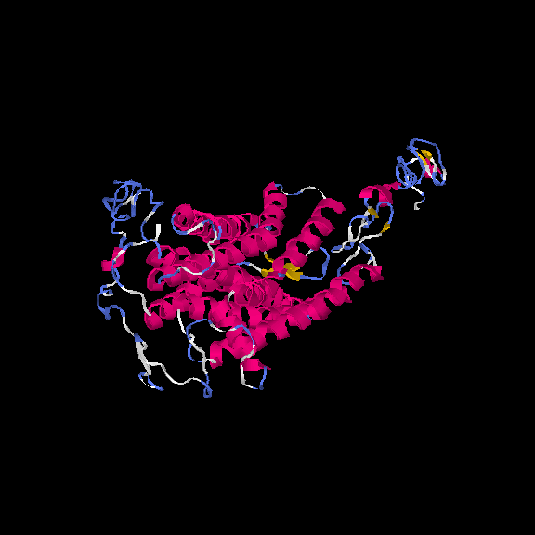 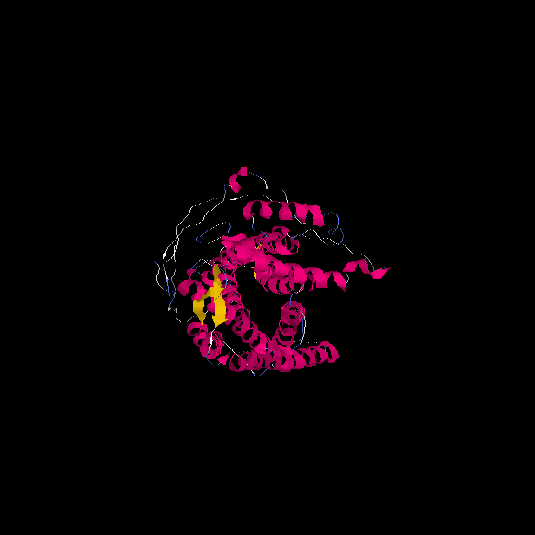  |
| HG0027 | P08173 | 479 | -0.4 | 0.66 ± 0.13 | 8.1 ± 4.4 | 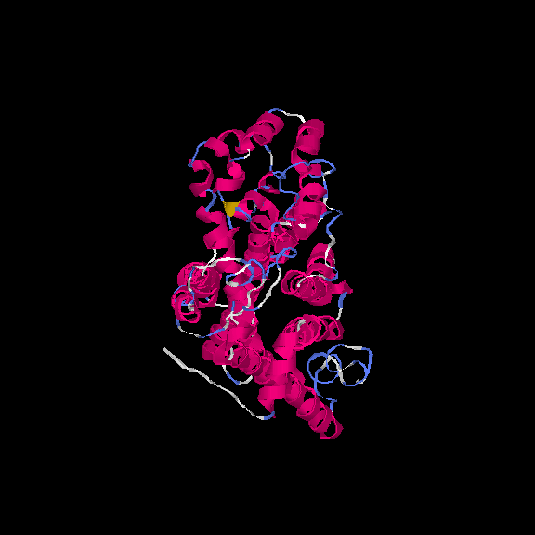 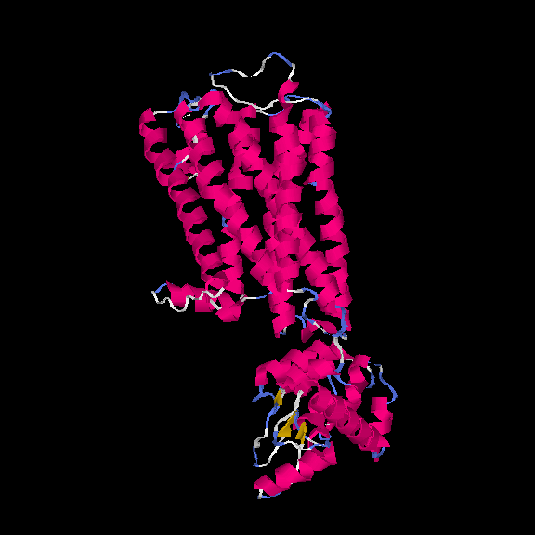 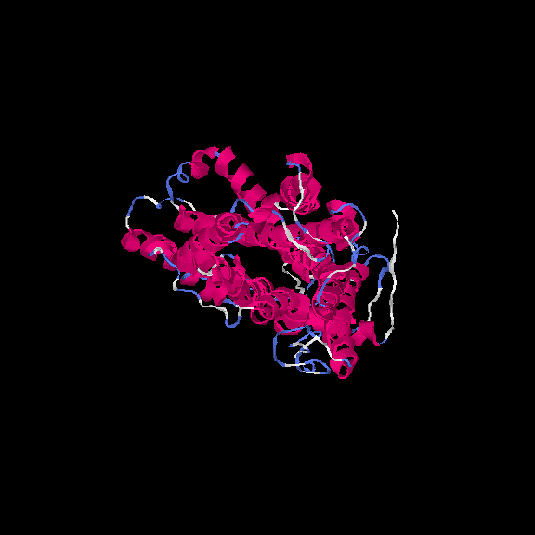 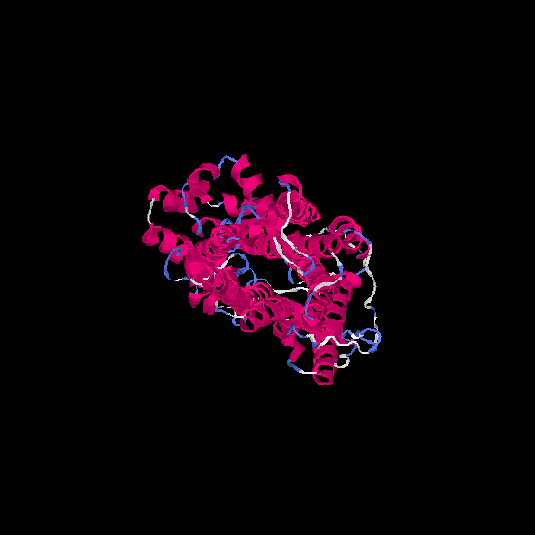  |
| HG0028 | Q5CZ62 | 431 | -1.65 | 0.51 ± 0.15 | 9.99 ± 4.6 |  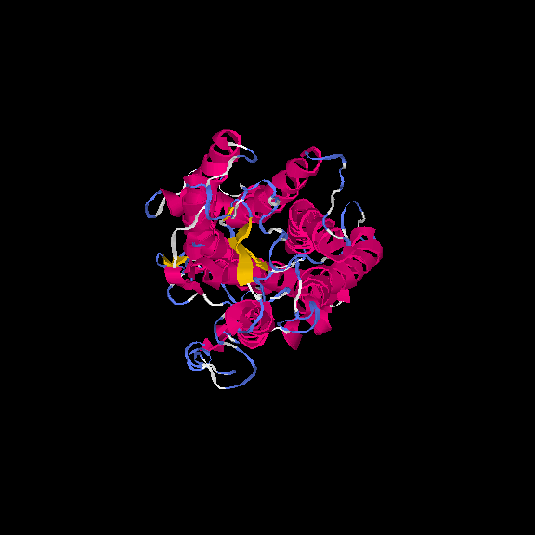 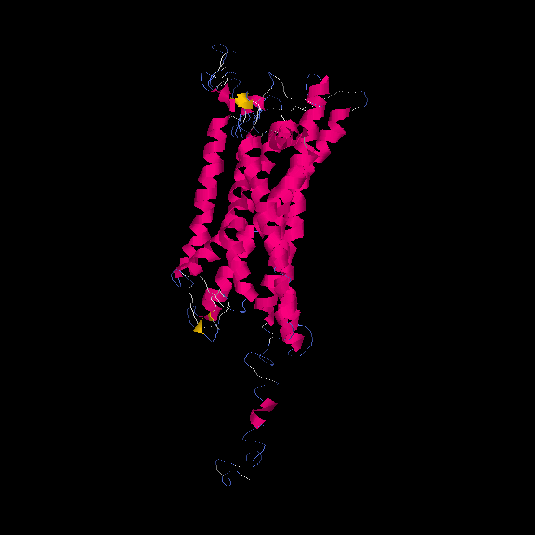 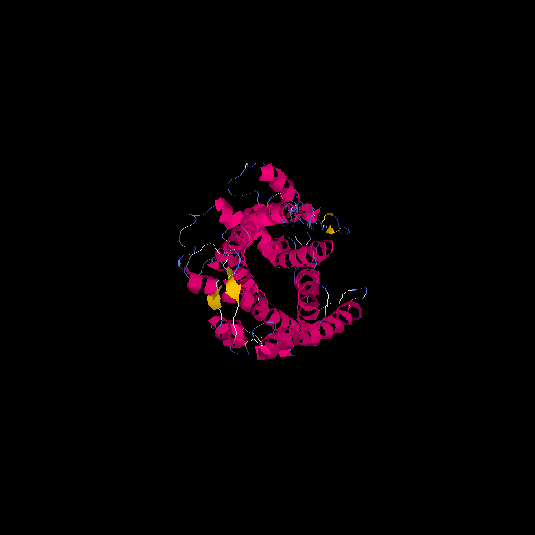 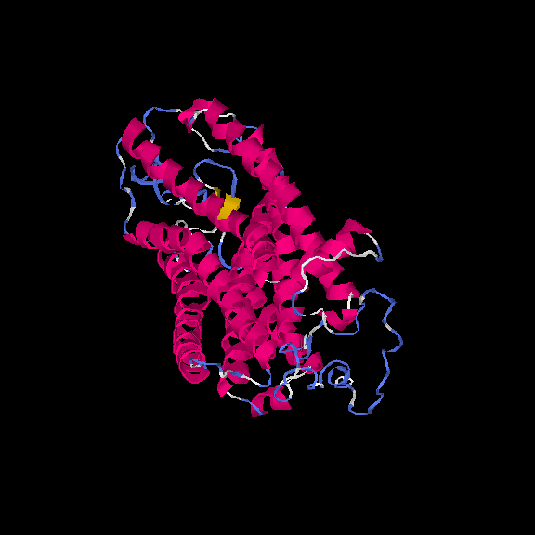 |
| HG0029 | P21462 | 350 | 0.02 | 0.72 ± 0.11 | 6.5 ± 3.9 | 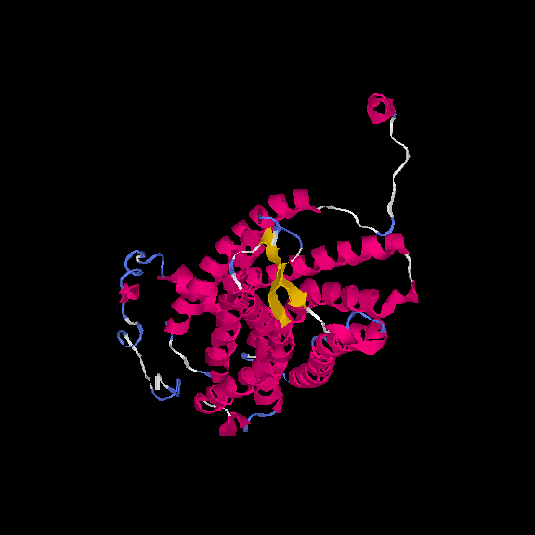 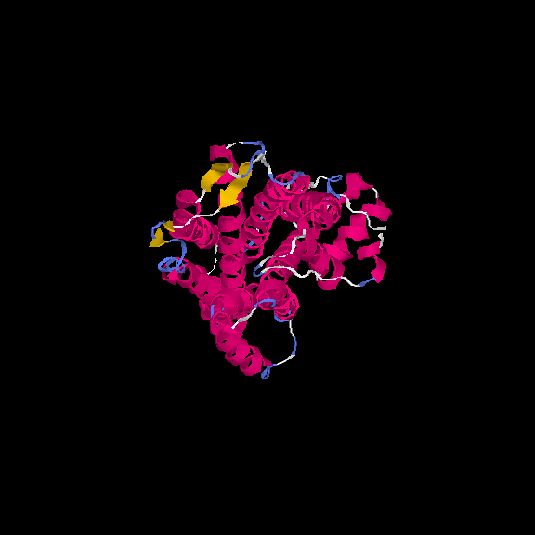 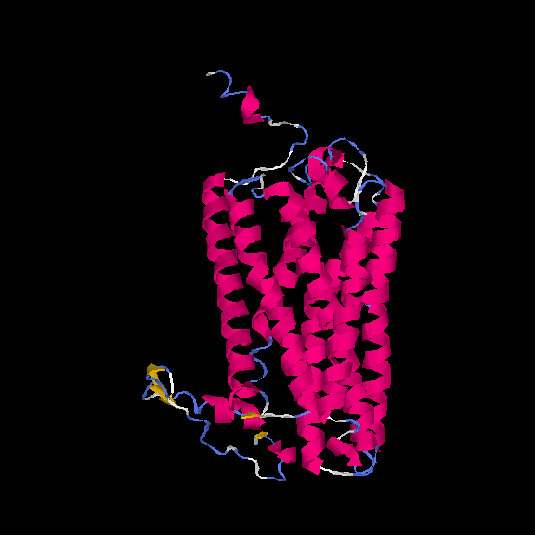 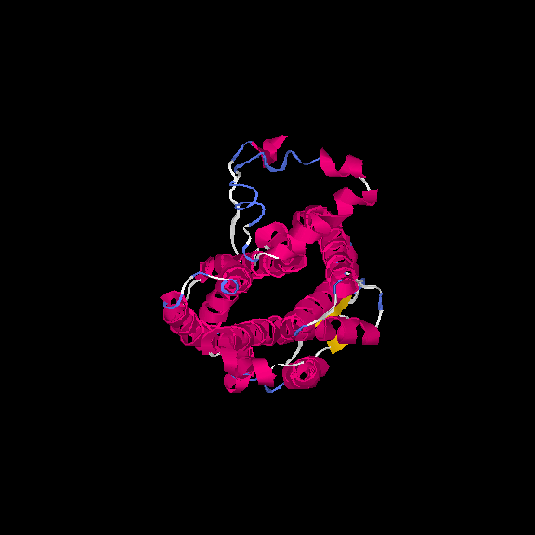 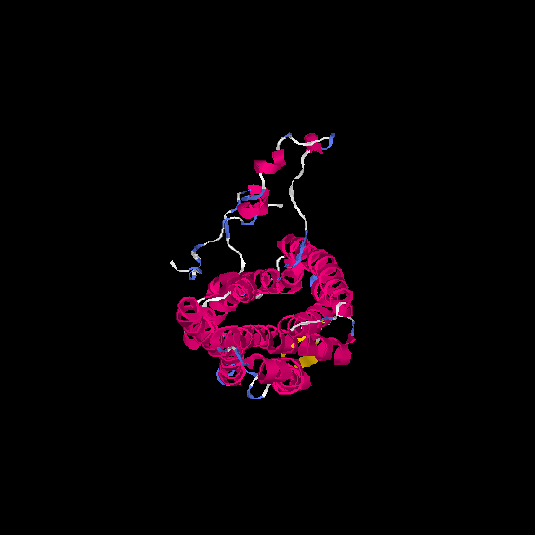 |
| HG0030 | P0C628 | 307 | -0.13 | 0.7 ± 0.12 | 6.5 ± 3.9 | 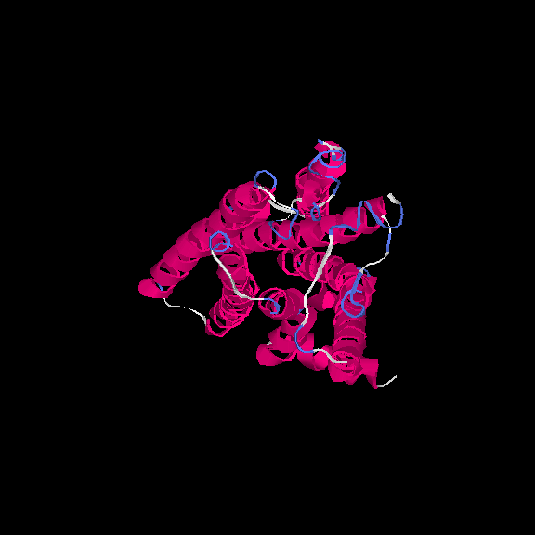 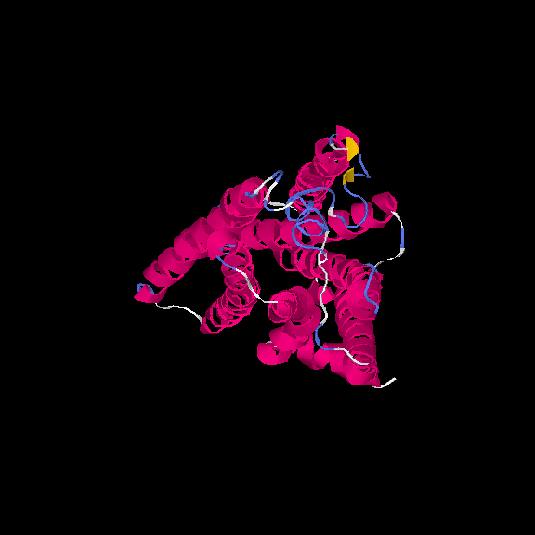 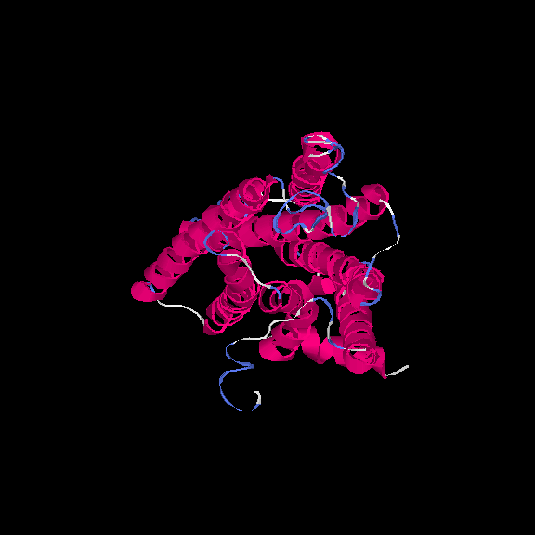 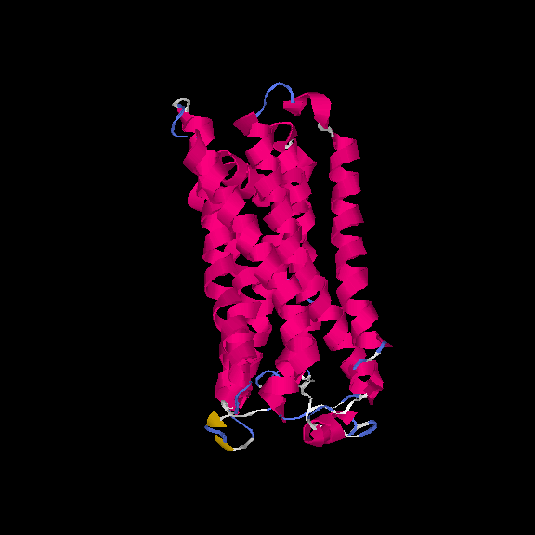 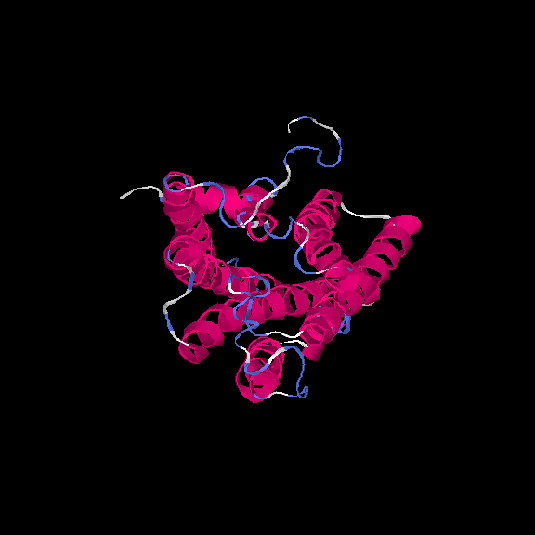 |
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218