


| HG ID | UniProt ID | Length | C-score | Estimated TM-score |
Estimated RMSD |
Top 5 models |
| HG0080 | Q6IF42 | 318 | -0.21 | 0.69 ± 0.12 | 6.8 ± 4 | 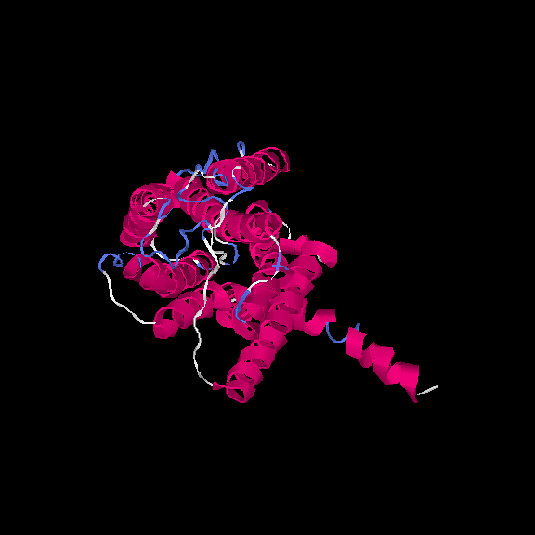  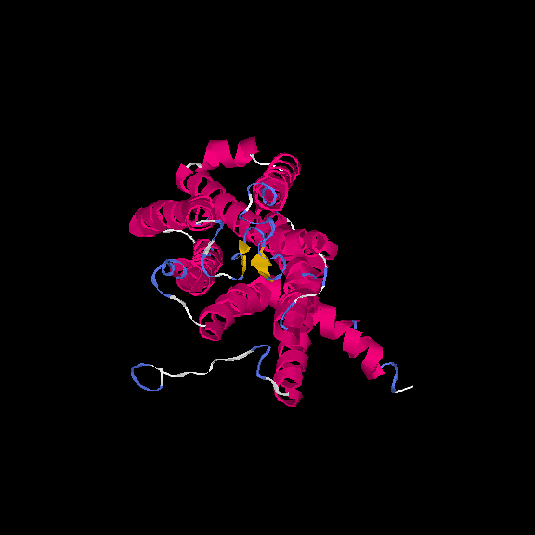  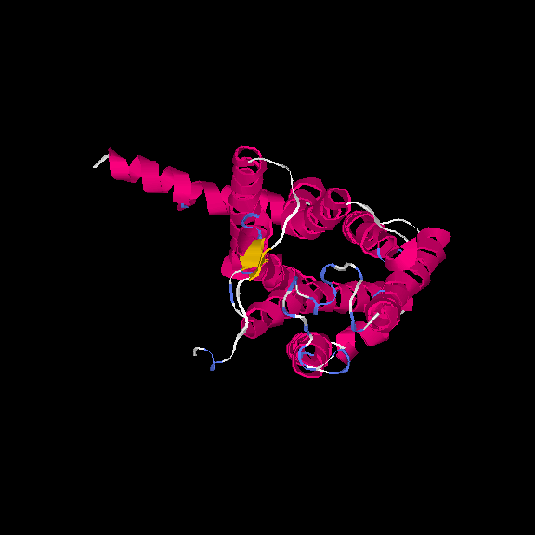 |
| HG0081 | Q8NGC9 | 324 | -0.58 | 0.64 ± 0.13 | 7.6 ± 4.3 | 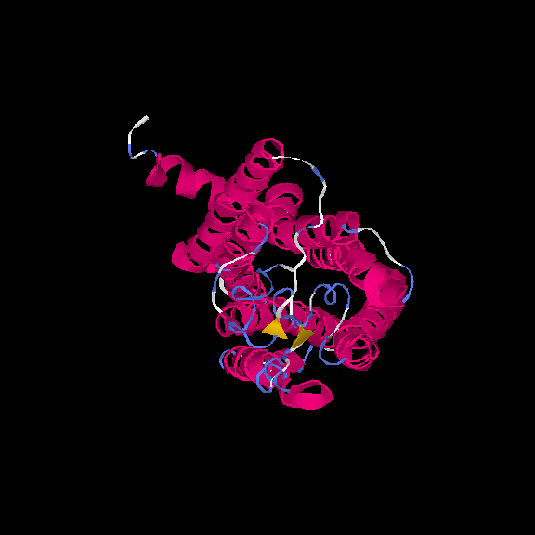 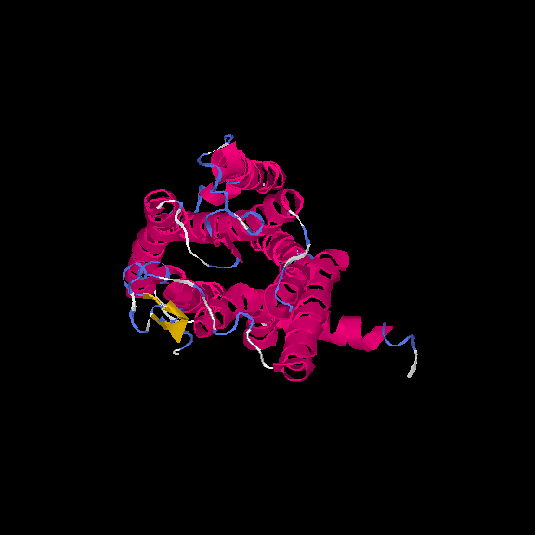 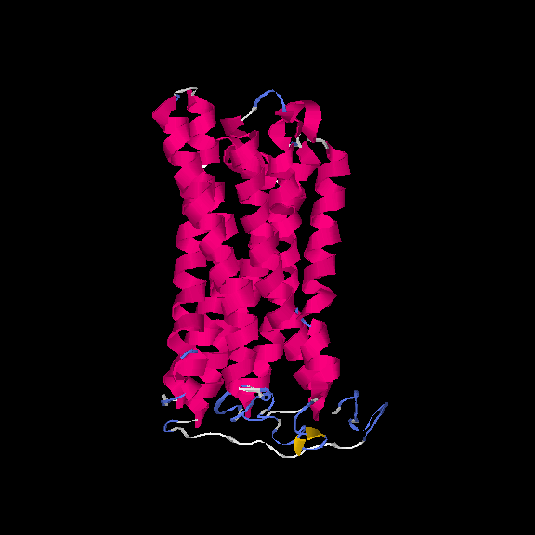 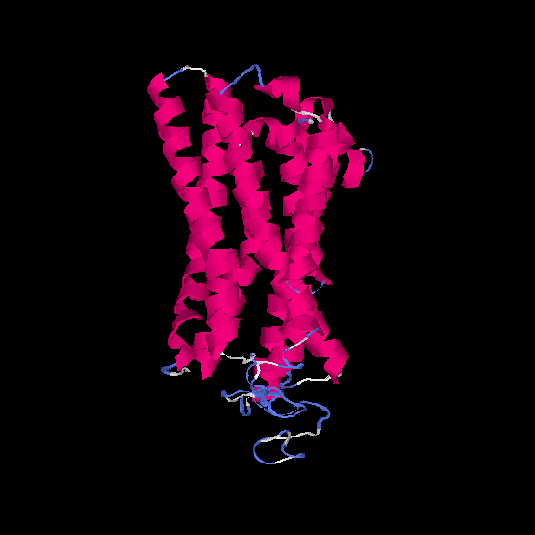 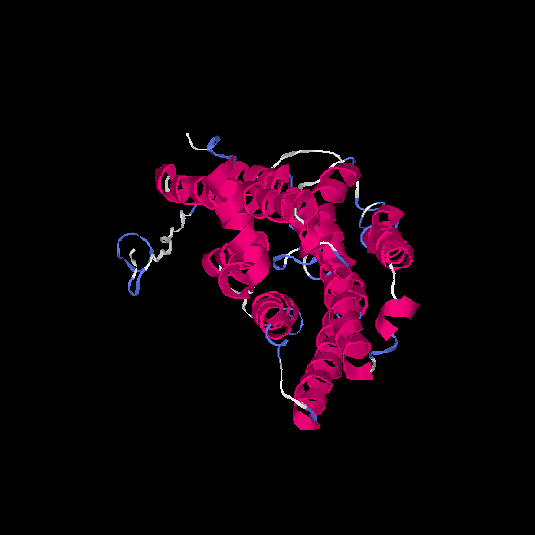 |
| HG0082 | Q8WZ84 | 308 | -0.02 | 0.71 ± 0.12 | 6.3 ± 3.8 | 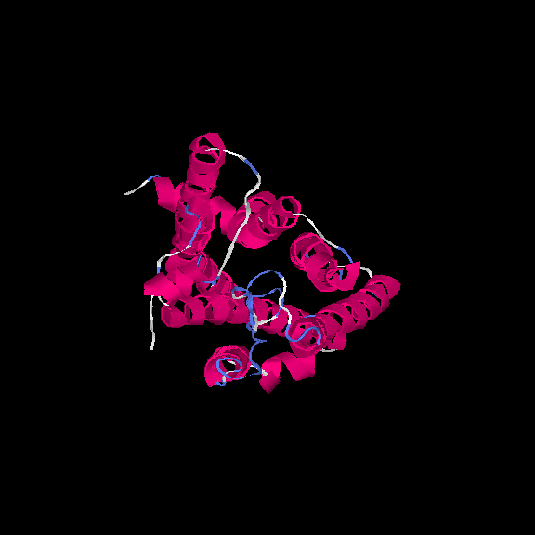  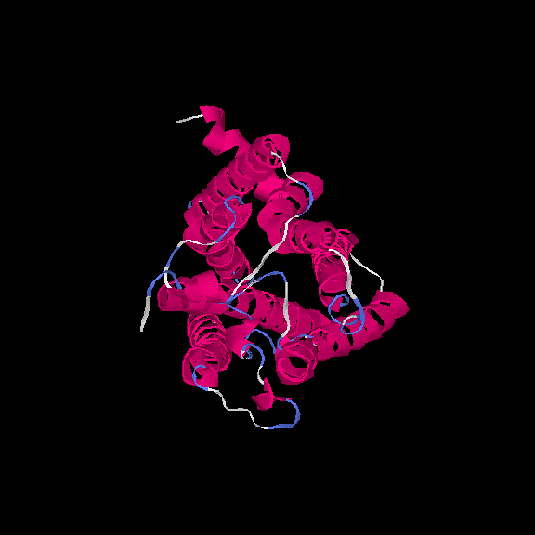 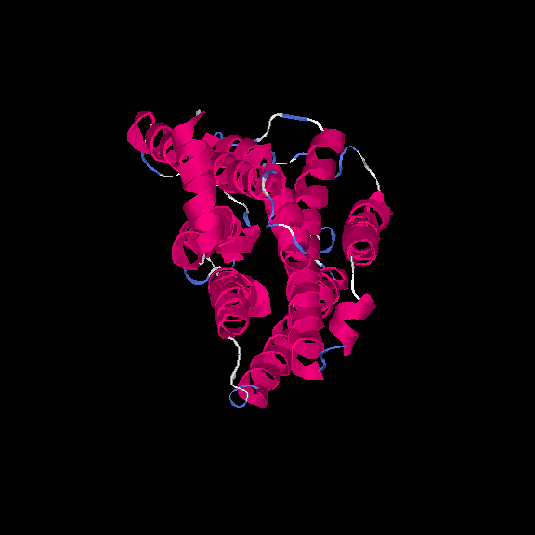 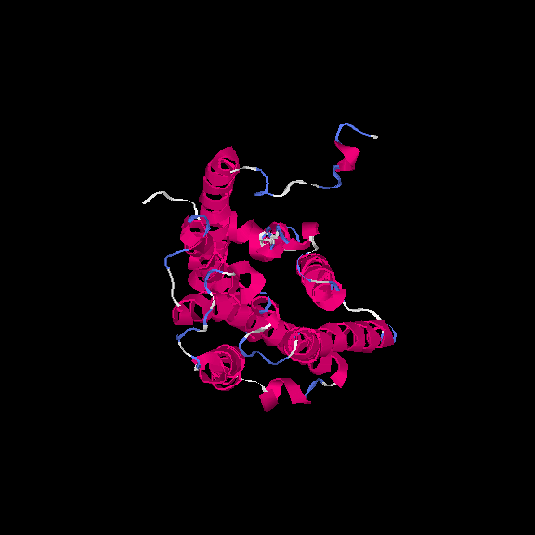 |
| HG0083 | Q8NH02 | 315 | -0.32 | 0.67 ± 0.13 | 7 ± 4.1 |     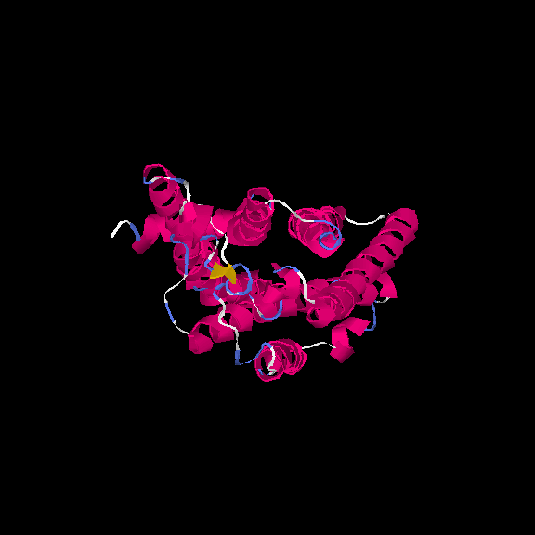 |
| HG0084 | O60412 | 319 | 0.01 | 0.72 ± 0.11 | 6.3 ± 3.8 |  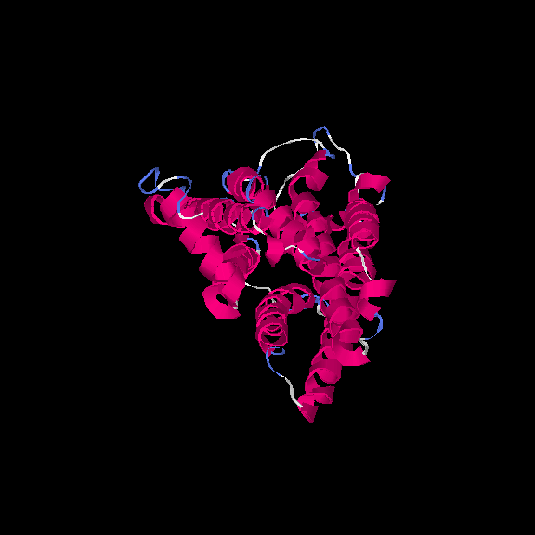 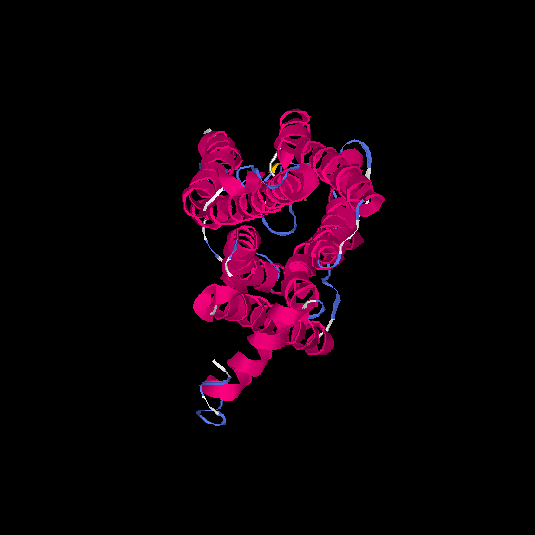 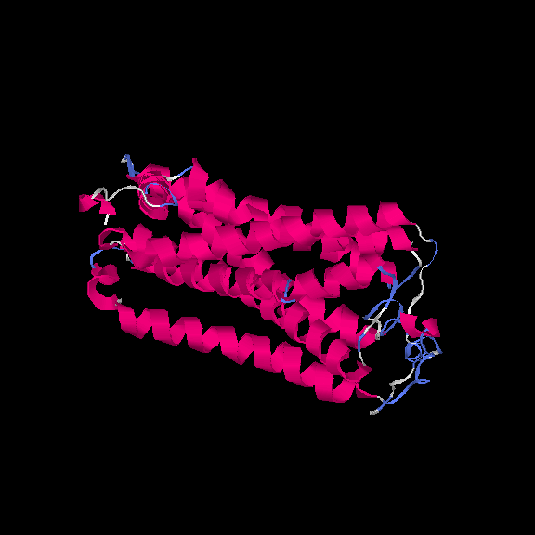 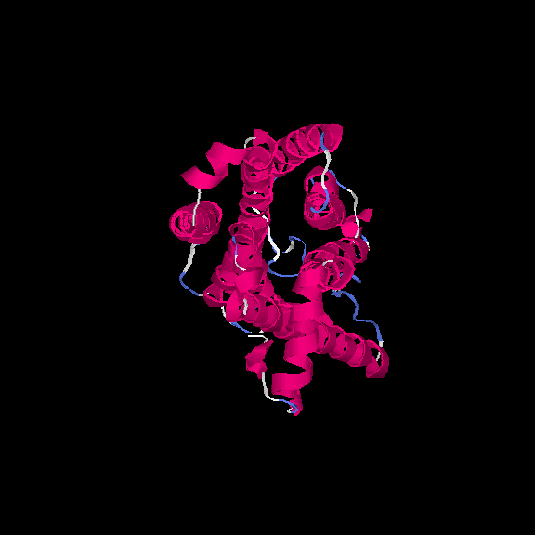 |
| HG0085 | Q9NYW2 | 309 | -0.06 | 0.71 ± 0.12 | 6.4 ± 3.9 | 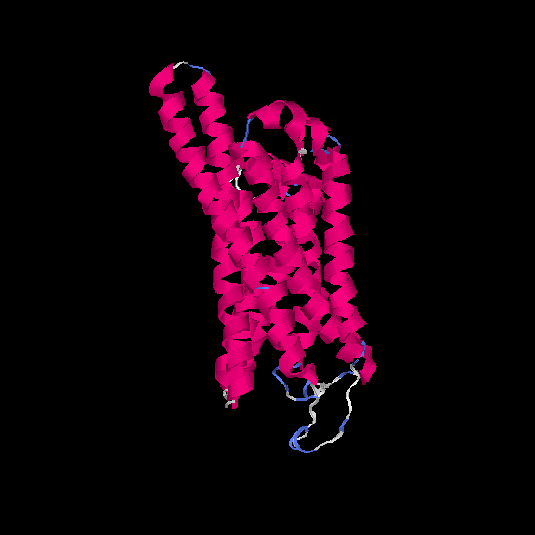 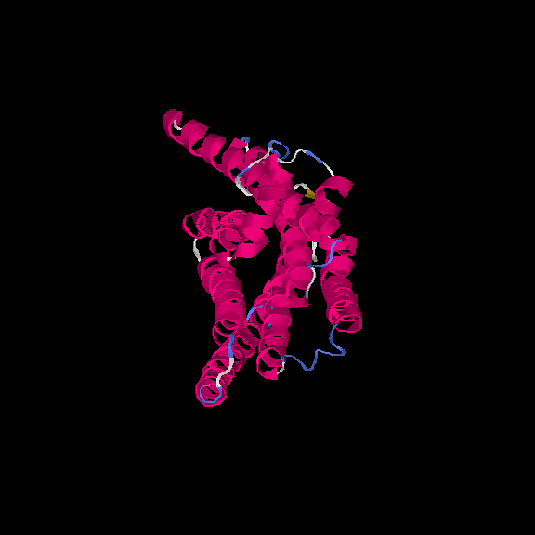 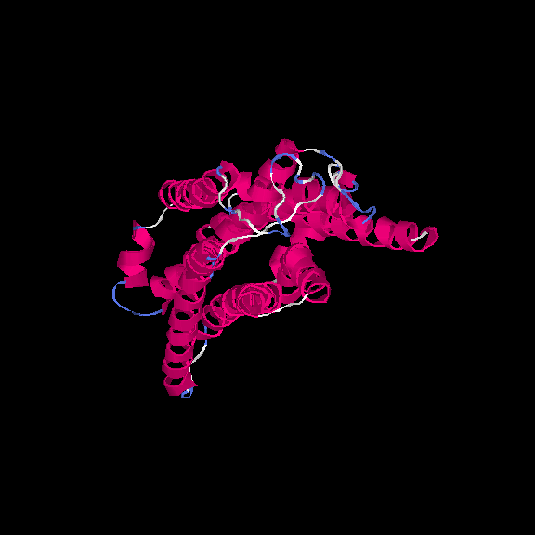 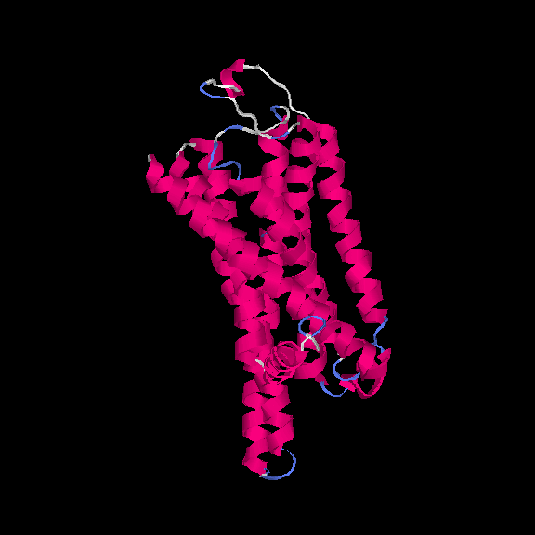 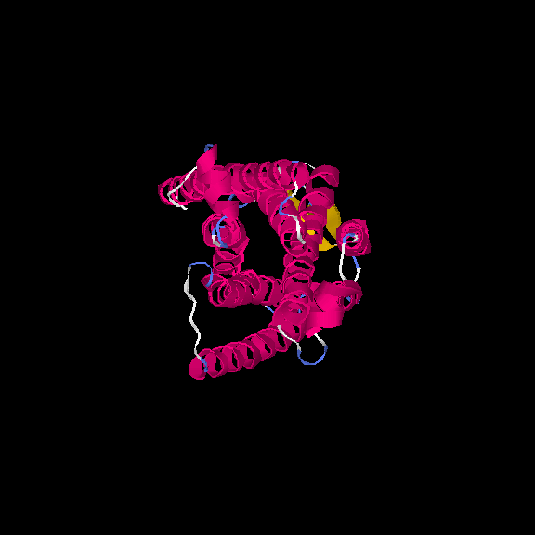 |
| HG0086 | P49685 | 360 | -0.66 | 0.63 ± 0.14 | 8 ± 4.4 | 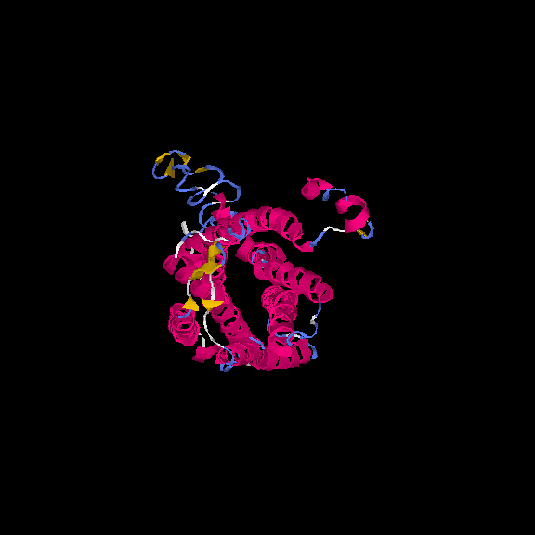  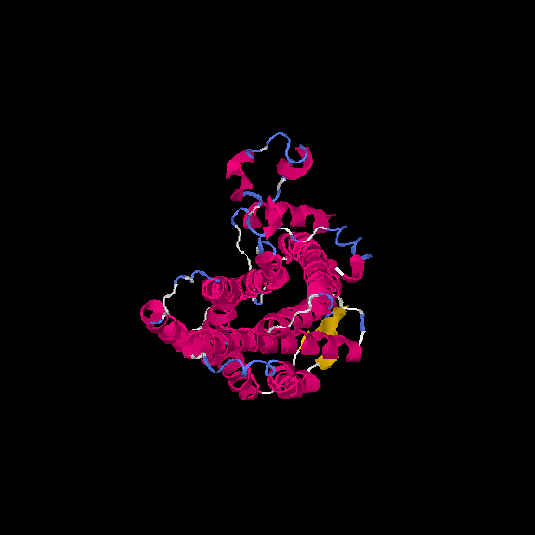 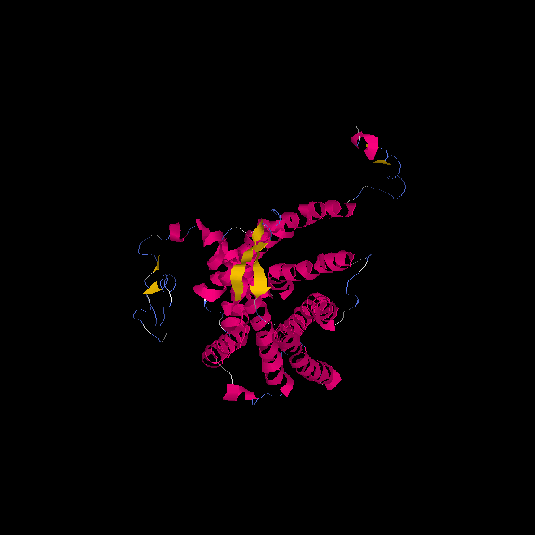 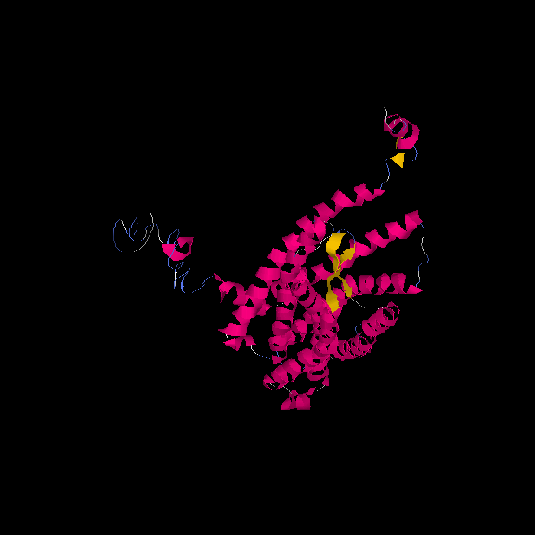 |
| HG0087 | Q8NGX8 | 325 | -0.39 | 0.66 ± 0.13 | 7.2 ± 4.2 | 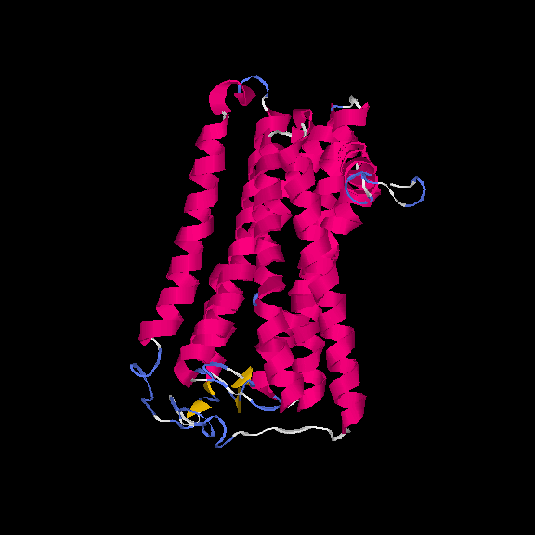 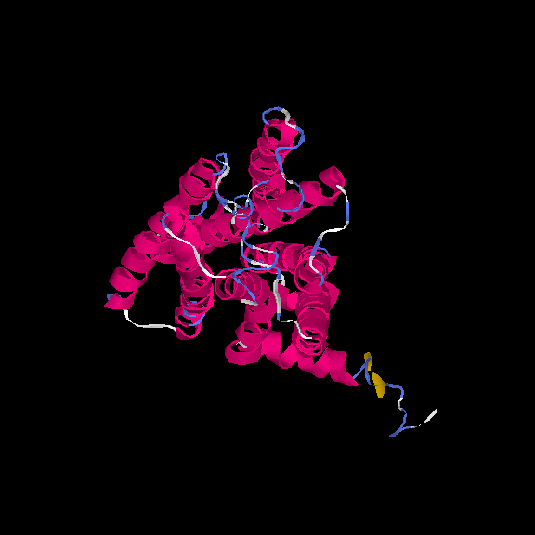    |
| HG0088 | Q4VBL8 | 406 | -1.4 | 0.54 ± 0.15 | 9.99 ± 4.6 |      |
| HG0089 | Q8NGZ0 | 328 | -0.15 | 0.69 ± 0.12 | 6.7 ± 4 | 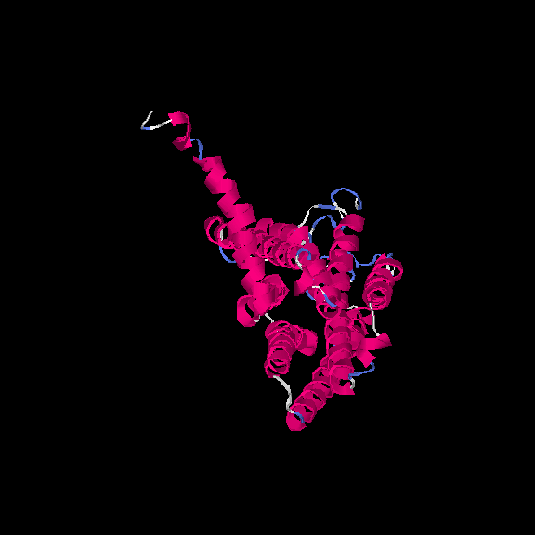 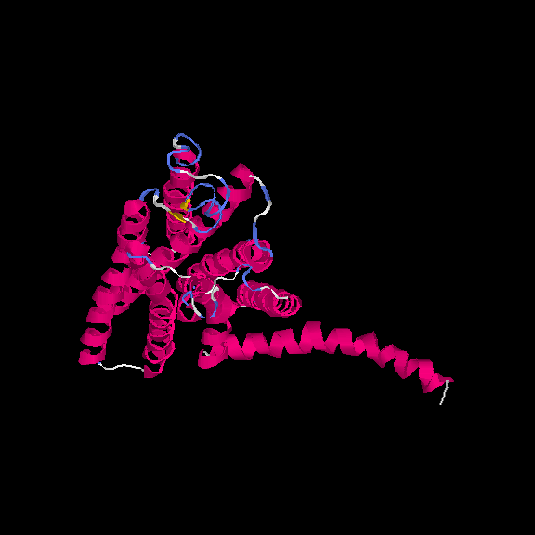 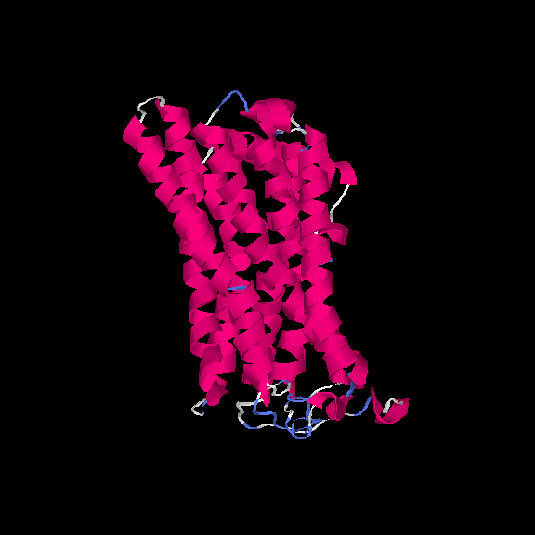  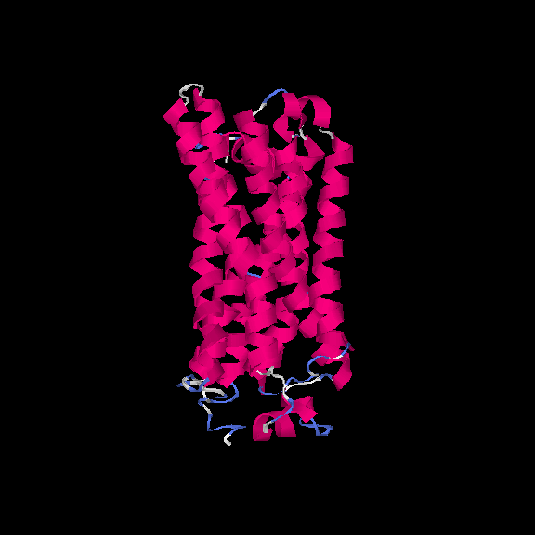 |
| HG0090 | Q8NGS3 | 322 | 0.1 | 0.73 ± 0.11 | 6.1 ± 3.8 | 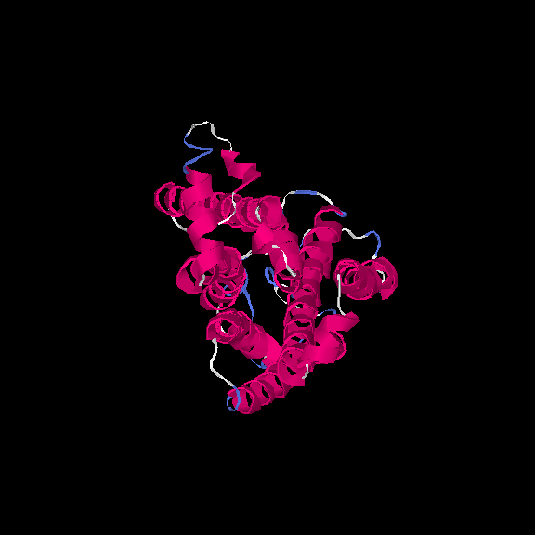 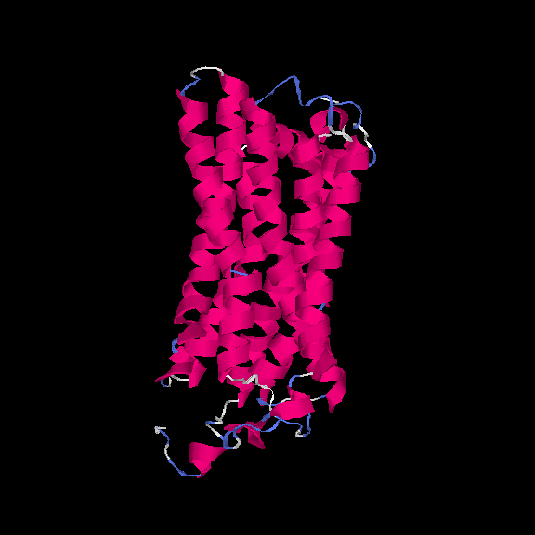 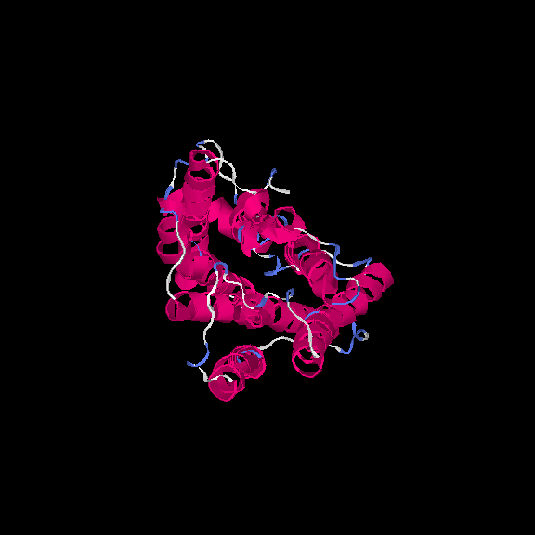 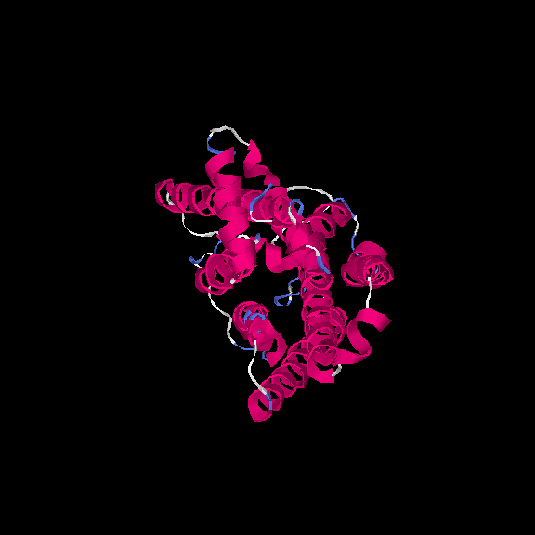  |
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218