

You can:
| Name | Mu-type opioid receptor |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Oprm1 |
| Synonym | Opioid receptor B opioid receptor OP3 mu receptor MOR-1 [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 398 |
| Amino acid sequence | MDSSTGPGNTSDCSDPLAQASCSPAPGSWLNLSHVDGNQSDPCGLNRTGLGGNDSLCPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGTILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCFREFCIPTSSTIEQQNSTRVRQNTREHPSTANTVDRTNHQLENLEAETAPLP |
| UniProt | P33535 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL270 |
| IUPHAR | 319 |
| DrugBank | N/A |
| Name | CHEMBL294616 |
|---|---|
| Molecular formula | C30H39N5O7S2 |
| IUPAC name | (4S,7R,13S)-13-[[(2S)-2-amino-3-(4-hydroxyphenyl)propanoyl]amino]-7-benzyl-3,3,14,14-tetramethyl-6,9,12-trioxo-1,2-dithia-5,8,11-triazacyclotetradecane-4-carboxylic acid |
| Molecular weight | 645.79 |
| Hydrogen bond acceptor | 10 |
| Hydrogen bond donor | 7 |
| XlogP | -0.6 |
| Synonyms | 13-[2-Amino-3-(4-hydroxy-phenyl)-propionylamino]-7-benzyl-3,3,14,14-tetramethyl-6,9,12-trioxo-1,2-dithia-5,8,11-triaza-cyclotetradecane-4-carboxylic acid BDBM50001683 MCMMCRYPQBNCPH-DVKRWUGUSA-N |
| Inchi Key | MCMMCRYPQBNCPH-DVKRWUGUSA-N |
| Inchi ID | InChI=1S/C30H39N5O7S2/c1-29(2)23(34-25(38)20(31)14-18-10-12-19(36)13-11-18)27(40)32-16-22(37)33-21(15-17-8-6-5-7-9-17)26(39)35-24(28(41)42)30(3,4)44-43-29/h5-13,20-21,23-24,36H,14-16,31H2,1-4H3,(H,32,40)(H,33,37)(H,34,38)(H,35,39)(H,41,42)/t20-,21+,23-,24-/m0/s1 |
| PubChem CID | 44299404 |
| ChEMBL | CHEMBL294616 |
| IUPHAR | N/A |
| BindingDB | 50001683 |
| DrugBank | N/A |
Structure | 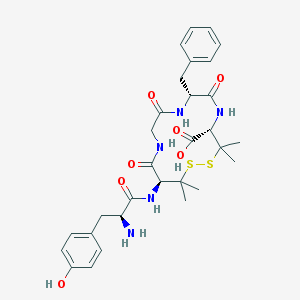 |
| Lipinski's druglikeness | This ligand has more than 5 hydrogen bond donor. This ligand is heavier than 500 daltons. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | <35000.0 nM | PMID8201592 | BindingDB |
| IC50 | 385.0 nM | PMID2537426 | BindingDB,ChEMBL |
| IC50 | 609.0 nM | PMID9822547, PMID2153205, PMID1320122 | BindingDB,ChEMBL |
| IC50 | 619.0 nM | PMID1648137 | BindingDB,ChEMBL |
| IC50 | 620.0 nM | PMID8201592, PMID8289188 | BindingDB,ChEMBL |
| IC50 | 1300.0 nM | PMID1851843 | BindingDB,ChEMBL |
| IC50 | 3700.0 nM | PMID9397176 | BindingDB,ChEMBL |
| Ki | 610.0 nM | PMID20560643, PMID20617791 | BindingDB,ChEMBL |
| Ki | 873.0 nM | PMID2845081 | BindingDB |
| Ki | 873.0 nM | PMID2845081, PMID2828622 | BindingDB,ChEMBL |
| Ki | 943.0 nM | PMID8230106 | BindingDB,ChEMBL |
| Ki | 2018.0 nM | PMID1311764 | BindingDB,ChEMBL |
| Ki | 2120.0 nM | PMID20599386 | BindingDB,ChEMBL |
| Ki | 2820.0 nM | PMID22325949 | ChEMBL |
| Potency | 0.01 - | PMID8230106 | ChEMBL |
| Ratio | 73.0 - | PMID8201592 | ChEMBL |
| Ratio | 116.0 - | PMID2153205 | ChEMBL |
| Ratio | 380.0 - | PMID1320122 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218