

You can:
| Name | Muscarinic acetylcholine receptor M2 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CHRM2 |
| Synonym | cholinergic receptor AChR M2 M2 muscarinic acetylcholine receptor M2 receptor Chrm-2 [ Show all ] |
| Disease | Urinary incontinence Heart failure Nausea; Addiction Parkinson's disease Peptic ulcer [ Show all ] |
| Length | 466 |
| Amino acid sequence | MNNSTNSSNNSLALTSPYKTFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVEDGECYIQFFSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRIKKDKKEPVANQDPVSPSLVQGRIVKPNNNNMPSSDDGLEHNKIQNGKAPRDPVTENCVQGEEKESSNDSTSVSAVASNMRDDEITQDENTVSTSLGHSKDENSKQTCIRIGTKTPKSDSCTPTNTTVEVVGSSGQNGDEKQNIVARKIVKMTKQPAKKKPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLMCHYKNIGATR |
| UniProt | P08172 |
| Protein Data Bank | 5zkc, 4mqs, 4mqt, 5yc8, 5zk3, 5zkb, 5zk8 |
| GPCR-HGmod model | P08172 |
| 3D structure model | This structure is from PDB ID 5zkc. |
| BioLiP | BL0433341, BL0433216, BL0263147, BL0263146, BL0263145, BL0433339, BL0433340, BL0433338 |
| Therapeutic Target Database | T46185 |
| ChEMBL | CHEMBL211 |
| IUPHAR | 14 |
| DrugBank | BE0000560 |
| Name | Methoctramine |
|---|---|
| Molecular formula | C36H62N4O2 |
| IUPAC name | N,N'-bis[6-[(2-methoxyphenyl)methylamino]hexyl]octane-1,8-diamine |
| Molecular weight | 582.918 |
| Hydrogen bond acceptor | 6 |
| Hydrogen bond donor | 4 |
| XlogP | 6.8 |
| Synonyms | CHEBI:73339 Lopac0_000862 N,N'-Bis[6-[[(2-methoxyphenyl)methyl]amino]hexyl]-1,8-octanediamine NCGC00162283-02 1,8-Octanediamine, N1,N8-bis(6-(((2-methoxyphenyl)methyl)amino)hexyl)- [ Show all ] |
| Inchi Key | RPMBYDYUVKEZJA-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C36H62N4O2/c1-41-35-23-13-11-21-33(35)31-39-29-19-9-7-17-27-37-25-15-5-3-4-6-16-26-38-28-18-8-10-20-30-40-32-34-22-12-14-24-36(34)42-2/h11-14,21-24,37-40H,3-10,15-20,25-32H2,1-2H3 |
| PubChem CID | 4108 |
| ChEMBL | CHEMBL27673 |
| IUPHAR | 327 |
| BindingDB | 50064176 |
| DrugBank | N/A |
Structure | 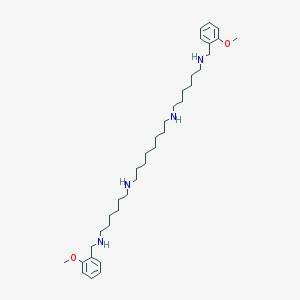 |
| Lipinski's druglikeness | This ligand is heavier than 500 daltons. This ligand has a partition coefficient log P greater than 5. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 2.2 nM | PMID26988801, PMID27876250 | BindingDB |
| IC50 | 34.0 nM | PMID20875743 | BindingDB |
| IC50 | 37.0 nM | PMID23466604 | ChEMBL |
| IC50 | 51.0 nM | PMID18983139 | ChEMBL |
| Kd | 12.02 nM | PMID9784091 | ChEMBL |
| Kd | 12.3 nM | PMID9767650 | ChEMBL |
| Kd | 15.14 nM | PMID7932564, PMID8246244 | BindingDB,ChEMBL |
| Kd | 19.5 nM | PMID2909747 | BindingDB,ChEMBL |
| Ki | 0.6 nM | PMID8496700 | BindingDB |
| Ki | 2.399 nM | PMID17276075 | ChEMBL |
| Ki | 3.6 nM | PMID2704370 | PDSP,BindingDB |
| Ki | 3.98108 - 50.1187 nM | PMID8759038, PMID2704370, PMID9454790, PMID16188951, PMID1994002, PMID9113359 | IUPHAR |
| Ki | 12.5893 nM | PMID8016895 | PDSP |
| Ki | 13.18 nM | PMID1994002 | BindingDB |
| Ki | 13.1826 nM | PMID1994002 | PDSP |
| Ki | 14.3 nM | PMID11708906 | BindingDB,ChEMBL |
| Ki | 14.35 nM | PMID7805774 | PDSP,BindingDB |
| Ki | 26.0 nM | PMID23466604 | ChEMBL |
| Ki | 27.0 nM | PMID24805037 | BindingDB,ChEMBL |
| Ki | 36.0 nM | PMID18983139 | ChEMBL |
| Ki | 42.0 nM | PMID9454790 | PDSP,BindingDB |
| Ki | 77.62 nM | PMID13679167 | BindingDB |
| Ki | 77.6247 nM | PMID13679167 | PDSP |
| pKb | 7.82 - | PMID17276075 | ChEMBL |
| pKB | 7.91 - | PMID12109912, PMID11960498 | ChEMBL |
| pKb | 7.92 - | PMID12620072 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218