

You can:
| Name | D(2) dopamine receptor |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Drd2 |
| Synonym | D2 receptor D2(415) and D2(444) D2A and D2B D2R Dopamine D2 receptor [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 444 |
| Amino acid sequence | MDPLNLSWYDDDLERQNWSRPFNGSEGKADRPHYNYYAMLLTLLIFIIVFGNVLVCMAVSREKALQTTTNYLIVSLAVADLLVATLVMPWVVYLEVVGEWKFSRIHCDIFVTLDVMMCTASILNLCAISIDRYTAVAMPMLYNTRYSSKRRVTVMIAIVWVLSFTISCPLLFGLNNTDQNECIIANPAFVVYSSIVSFYVPFIVTLLVYIKIYIVLRKRRKRVNTKRSSRAFRANLKTPLKGNCTHPEDMKLCTVIMKSNGSFPVNRRRMDAARRAQELEMEMLSSTSPPERTRYSPIPPSHHQLTLPDPSHHGLHSNPDSPAKPEKNGHAKIVNPRIAKFFEIQTMPNGKTRTSLKTMSRRKLSQQKEKKATQMLAIVLGVFIICWLPFFITHILNIHCDCNIPPVLYSAFTWLGYVNSAVNPIIYTTFNIEFRKAFMKILHC |
| UniProt | P61169 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL339 |
| IUPHAR | 215 |
| DrugBank | N/A |
| Name | n-Propylapomorphine |
|---|---|
| Molecular formula | C19H21NO2 |
| IUPAC name | (6aR)-6-propyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,11-diol |
| Molecular weight | 295.382 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 2 |
| XlogP | 3.2 |
| Synonyms | BDBM50012994 CHEMBL538542 PDSP1_000853 SKF-76783 (9R)-10-propyl-10-azatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(17),2,4,6,13,15-hexaene-3,4-diol [ Show all ] |
| Inchi Key | BTGAJCKRXPNBFI-OAHLLOKOSA-N |
| Inchi ID | InChI=1S/C19H21NO2/c1-2-9-20-10-8-12-4-3-5-14-17(12)15(20)11-13-6-7-16(21)19(22)18(13)14/h3-7,15,21-22H,2,8-11H2,1H3/t15-/m1/s1 |
| PubChem CID | 167715 |
| ChEMBL | CHEMBL225230 |
| IUPHAR | 969 |
| BindingDB | 50012994, 50007422 |
| DrugBank | N/A |
Structure | 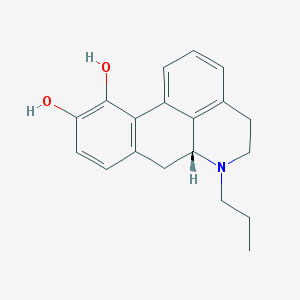 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 4.8 nM | PMID2147956 | BindingDB,ChEMBL |
| IC50 | 10.0 nM | PMID2968456 | BindingDB,ChEMBL |
| Kd | 9.0 nM | PMID10956209 | BindingDB,ChEMBL |
| Kd | 40.0 nM | PMID10956209 | BindingDB,ChEMBL |
| Kd | 370.0 nM | PMID10956209 | BindingDB,ChEMBL |
| Kd | 520.0 nM | PMID10956209 | BindingDB,ChEMBL |
| Ki | <10000.0 nM | PMID9021894 | BindingDB |
| Ki | 0.18 nM | PMID21666830 | BindingDB,ChEMBL |
| Ki | 0.8 nM | PMID2136919, PMID1671415 | BindingDB |
| Ki | 0.8 nM | PMID1971309, PMID1671415 | BindingDB,ChEMBL |
| Ki | 0.9 nM | PMID18251489 | ChEMBL |
| Ki | 10.0 nM | PMID17228858 | BindingDB,ChEMBL |
| Ki | 54.0 nM | PMID21666830 | BindingDB,ChEMBL |
| Ki | 460.0 nM | PMID9021894 | BindingDB |
| KL | 23.0 nM | PMID8632409 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218