

You can:
| Name | Histamine H3 receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HRH3 |
| Synonym | HH3R H3R H3 receptor GPCR97 G-protein coupled receptor 97 |
| Disease | Obese insulin-resistant disorders Excessive daytime sleepiness Sleep disorders Schizophrenia Pain [ Show all ] |
| Length | 445 |
| Amino acid sequence | MERAPPDGPLNASGALAGEAAAAGGARGFSAAWTAVLAALMALLIVATVLGNALVMLAFVADSSLRTQNNFFLLNLAISDFLVGAFCIPLYVPYVLTGRWTFGRGLCKLWLVVDYLLCTSSAFNIVLISYDRFLSVTRAVSYRAQQGDTRRAVRKMLLVWVLAFLLYGPAILSWEYLSGGSSIPEGHCYAEFFYNWYFLITASTLEFFTPFLSVTFFNLSIYLNIQRRTRLRLDGAREAAGPEPPPEAQPSPPPPPGCWGCWQKGHGEAMPLHRYGVGEAAVGAEAGEATLGGGGGGGSVASPTSSSGSSSRGTERPRSLKRGSKPSASSASLEKRMKMVSQSFTQRFRLSRDRKVAKSLAVIVSIFGLCWAPYTLLMIIRAACHGHCVPDYWYETSFWLLWANSAVNPVLYPLCHHSFRRAFTKLLCPQKLKIQPHSSLEHCWK |
| UniProt | Q9Y5N1 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q9Y5N1 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q9Y5N1. |
| BioLiP | N/A |
| Therapeutic Target Database | T64765 |
| ChEMBL | CHEMBL264 |
| IUPHAR | 264 |
| DrugBank | BE0000968 |
| Name | clobenpropit |
|---|---|
| Molecular formula | C14H17ClN4S |
| IUPAC name | 3-(1H-imidazol-5-yl)propyl N'-[(4-chlorophenyl)methyl]carbamimidothioate |
| Molecular weight | 308.828 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 2 |
| XlogP | 3.0 |
| Synonyms | BDBM50214615 HMS3394B07 N'-[(4-chlorophenyl)methyl]-1-[3-(3H-imidazol-4-yl)propylsulfanyl]methanimidamide SAM001247107 Vuf-9153 [ Show all ] |
| Inchi Key | UCAIEVHKDLMIFL-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C14H17ClN4S/c15-12-5-3-11(4-6-12)8-18-14(16)20-7-1-2-13-9-17-10-19-13/h3-6,9-10H,1-2,7-8H2,(H2,16,18)(H,17,19) |
| PubChem CID | 2790 |
| ChEMBL | CHEMBL14690 |
| IUPHAR | 1223 |
| BindingDB | 50214615, 22541 |
| DrugBank | N/A |
Structure | 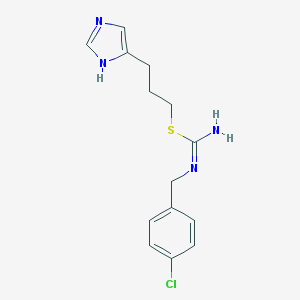 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | -62.8 - | PMID18433114 | ChEMBL |
| IC50 | 0.7943 nM | PMID24140447 | ChEMBL |
| IC50 | 1.0 nM | PMID20833043 | BindingDB,ChEMBL |
| Kd | 0.1 nM | PMID24140447 | ChEMBL |
| Ki | <10000.0 nM | PMID10347254 | PDSP,BindingDB |
| Ki | 0.3388 nM | PMID18433114 | ChEMBL |
| Ki | 0.339 nM | PMID18433114 | BindingDB |
| Ki | 0.36 nM | PMID12606603 | BindingDB |
| Ki | 0.363078 nM | PMID12606603 | PDSP |
| Ki | 0.39 - 3.9 nM | PMID12606603, PMID10869375, PMID11284713, PMID11714875, PMID11090094, PMID12393057, PMID11179434 | IUPHAR |
| Ki | 0.58 nM | PMID15139761 | BindingDB,ChEMBL |
| Ki | 0.6 nM | PMID10869375, PMID11179434, PMID15634025 | PDSP,BindingDB,ChEMBL |
| Ki | 0.776247 nM | PMID12606603 | PDSP |
| Ki | 0.78 nM | PMID12606603 | BindingDB |
| Ki | 2.4 nM | PMID19846299, PMID12672253, PMID21498080, PMID11294398 | BindingDB,ChEMBL |
| Ki | 2.818 nM | PMID22003888 | ChEMBL |
| Ki | 2.82 nM | PMID22003888 | BindingDB |
| Ki | 3.0 nM | PMID15947036 | BindingDB |
| Ki | 3.16 nM | PMID19414267 | BindingDB |
| Ki | 3.162 nM | PMID19414267 | ChEMBL |
| Ki | 3.8 nM | PMID11714875 | BindingDB |
| Ki | 3.80189 nM | PMID11714875 | PDSP |
| Ratio | 2.0 - | PMID11294398 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218