

You can:
| Name | Metabotropic glutamate receptor 2 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | GRM2 |
| Synonym | mGluR2 mGlu2 receptor metabotropic glutamate receptor 2 GPRC1B glutamate receptor |
| Disease | Central nervous system disease Anxiety disorder Bipolar disorder Major depressive disorder Mood disorder [ Show all ] |
| Length | 872 |
| Amino acid sequence | MGSLLALLALLLLWGAVAEGPAKKVLTLEGDLVLGGLFPVHQKGGPAEDCGPVNEHRGIQRLEAMLFALDRINRDPHLLPGVRLGAHILDSCSKDTHALEQALDFVRASLSRGADGSRHICPDGSYATHGDAPTAITGVIGGSYSDVSIQVANLLRLFQIPQISYASTSAKLSDKSRYDYFARTVPPDFFQAKAMAEILRFFNWTYVSTVASEGDYGETGIEAFELEARARNICVATSEKVGRAMSRAAFEGVVRALLQKPSARVAVLFTRSEDARELLAASQRLNASFTWVASDGWGALESVVAGSEGAAEGAITIELASYPISDFASYFQSLDPWNNSRNPWFREFWEQRFRCSFRQRDCAAHSLRAVPFEQESKIMFVVNAVYAMAHALHNMHRALCPNTTRLCDAMRPVNGRRLYKDFVLNVKFDAPFRPADTHNEVRFDRFGDGIGRYNIFTYLRAGSGRYRYQKVGYWAEGLTLDTSLIPWASPSAGPLPASRCSEPCLQNEVKSVQPGEVCCWLCIPCQPYEYRLDEFTCADCGLGYWPNASLTGCFELPQEYIRWGDAWAVGPVTIACLGALATLFVLGVFVRHNATPVVKASGRELCYILLGGVFLCYCMTFIFIAKPSTAVCTLRRLGLGTAFSVCYSALLTKTNRIARIFGGAREGAQRPRFISPASQVAICLALISGQLLIVVAWLVVEAPGTGKETAPERREVVTLRCNHRDASMLGSLAYNVLLIALCTLYAFKTRKCPENFNEAKFIGFTMYTTCIIWLAFLPIFYVTSSDYRVQTTTMCVSVSLSGSVVLGCLFAPKLHIILFQPQKNVVSHRAPTSRFGSAAARASSSLGQGSGSQFVPTVCNGREVVDSTTSSL |
| UniProt | Q14416 |
| Protein Data Bank | 5cnj, 5kzn, 5kzq, 5cni, 4xas, 4xaq |
| GPCR-HGmod model | N/A |
| 3D structure model | This structure is from PDB ID 5cnj. |
| BioLiP | BL0365442, BL0324559,BL0324560, BL0324557,BL0324558, BL0306697,BL0306698, BL0306695,BL0306696, BL0365443 |
| Therapeutic Target Database | T62820 |
| ChEMBL | CHEMBL5137 |
| IUPHAR | 290 |
| DrugBank | N/A |
| Name | SCHEMBL11235794 |
|---|---|
| Molecular formula | C5H9NO4 |
| IUPAC name | (2S)-2-amino-5-hydroxy-5-oxopentanoate;hydron |
| Molecular weight | 147.13 |
| Hydrogen bond acceptor | 5 |
| Hydrogen bond donor | 3 |
| XlogP | None |
| Synonyms | N/A |
| Inchi Key | WHUUTDBJXJRKMK-VKHMYHEASA-N |
| Inchi ID | InChI=1S/C5H9NO4/c6-3(5(9)10)1-2-4(7)8/h3H,1-2,6H2,(H,7,8)(H,9,10)/t3-/m0/s1 |
| PubChem CID | 88747398 |
| ChEMBL | CHEMBL575060 |
| IUPHAR | 1369 |
| BindingDB | 17657 |
| DrugBank | DB00142 |
Structure | 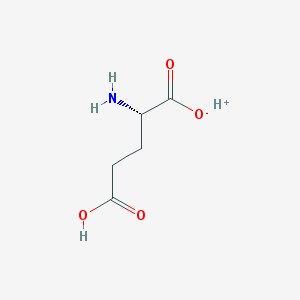 |
| Lipinski's druglikeness | Partition coefficient log P of this ligand is not available. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | 4000.0 nM | PMID12166935 | ChEMBL |
| EC50 | 290.0 nM | PMID9572889 | ChEMBL |
| EC50 | 1767.0 nM | PMID26313429 | BindingDB,ChEMBL |
| EC50 | 1800.0 nM | MedChemComm, (2011) 2:11:1120, PMID26814576 | BindingDB,ChEMBL |
| EC50 | 1820.0 nM | PMID26814576 | BindingDB |
| EC50 | 1827.0 nM | PMID26313429 | BindingDB,ChEMBL |
| EC50 | 2351.0 nM | PMID26313429 | BindingDB,ChEMBL |
| EC50 | 2575.0 nM | PMID26313429 | BindingDB,ChEMBL |
| EC50 | 3981.07 - 19952.6 nM | PMID10443583 | IUPHAR |
| EC50 | 4700.0 nM | PMID7738999 | BindingDB,ChEMBL |
| EC50 | 7400.0 nM | PMID11720869 | BindingDB,ChEMBL |
| EC50 | 8500.0 nM | PMID8759641 | BindingDB,ChEMBL |
| EC50 | 11800.0 nM | PMID7738999 | BindingDB |
| EC50 | 12589.2 nM | PMID21232953 | ChEMBL |
| EC50 | 13080.0 nM | PMID26814576 | BindingDB |
| EC50 | 16090.0 nM | PMID26814576 | BindingDB |
| EC50 | 26000.0 nM | PMID16213710, PMID17350267 | BindingDB,ChEMBL |
| Emax | 88.0 % | PMID26313429 | ChEMBL |
| Emax | 91.0 % | PMID26313429 | ChEMBL |
| Emax | 93.0 % | PMID26313429 | ChEMBL |
| Emax | 94.0 % | PMID26313429 | ChEMBL |
| Ki | <10000.0 nM | PMID8863838 | BindingDB |
| Ki | 1100.0 nM | PMID9463480 | BindingDB |
| Ki | 1200.0 nM | PMID12109902 | ChEMBL |
| Ki | 1258.93 nM | PMID12649361 | BindingDB |
| Ki | 1819.7 nM | MedChemComm, (2011) 2:11:1120 | ChEMBL |
| Ki | 4000.0 nM | PMID9131252 | BindingDB |
| Ki | 4400.0 nM | PMID9696422 | BindingDB |
| Ki | 6400.0 nM | PMID19042134 | BindingDB,ChEMBL |
| Ki | 11000.0 nM | PMID17725337 | BindingDB,ChEMBL |
| Ki | 12000.0 nM | PMID10893301 | BindingDB,ChEMBL |
| Ki | 12589.3 nM | PMID10530814 | IUPHAR |
| Ratio | 5.9 - | PMID12109902 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218