

You can:
| Name | Muscarinic acetylcholine receptor M4 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CHRM4 |
| Synonym | HM3 M4 receptor cholinergic receptor Chrm-4 cholinergic receptor, muscarinic 4 |
| Disease | Produce mydriasis and cycloplegia for diagnostic purposes Hypertension Irritable bowel syndrome Moderate and severe psychomotor agitation Mydriasis diagnosis [ Show all ] |
| Length | 479 |
| Amino acid sequence | MANFTPVNGSSGNQSVRLVTSSSHNRYETVEMVFIATVTGSLSLVTVVGNILVMLSIKVNRQLQTVNNYFLFSLACADLIIGAFSMNLYTVYIIKGYWPLGAVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPARRTTKMAGLMIAAAWVLSFVLWAPAILFWQFVVGKRTVPDNQCFIQFLSNPAVTFGTAIAAFYLPVVIMTVLYIHISLASRSRVHKHRPEGPKEKKAKTLAFLKSPLMKQSVKKPPPGEAAREELRNGKLEEAPPPALPPPPRPVADKDTSNESSSGSATQNTKERPATELSTTEATTPAMPAPPLQPRALNPASRWSKIQIVTKQTGNECVTAIEIVPATPAGMRPAANVARKFASIARNQVRKKRQMAARERKVTRTIFAILLAFILTWTPYNVMVLVNTFCQSCIPDTVWSIGYWLCYVNSTINPACYALCNATFKKTFRHLLLCQYRNIGTAR |
| UniProt | P08173 |
| Protein Data Bank | 5dsg |
| GPCR-HGmod model | P08173 |
| 3D structure model | This structure is from PDB ID 5dsg. |
| BioLiP | BL0339919,BL0339921, BL0339920 |
| Therapeutic Target Database | T20709, T50918 |
| ChEMBL | CHEMBL1821 |
| IUPHAR | 16 |
| DrugBank | BE0000405 |
| Name | Pecazine |
|---|---|
| Molecular formula | C19H22N2S |
| IUPAC name | 10-[(1-methylpiperidin-3-yl)methyl]phenothiazine |
| Molecular weight | 310.459 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 0 |
| XlogP | 5.6 |
| Synonyms | 10H-Phenothiazine, 10-[(1-methyl-3-piperidinyl)methyl]- AKOS021820849 CHEBI:135324 EINECS 200-490-8 MEPAZINE [ Show all ] |
| Inchi Key | CBHCDHNUZWWAPP-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C19H22N2S/c1-20-12-6-7-15(13-20)14-21-16-8-2-4-10-18(16)22-19-11-5-3-9-17(19)21/h2-5,8-11,15H,6-7,12-14H2,1H3 |
| PubChem CID | 6075 |
| ChEMBL | CHEMBL395110 |
| IUPHAR | N/A |
| BindingDB | N/A |
| DrugBank | N/A |
Structure | 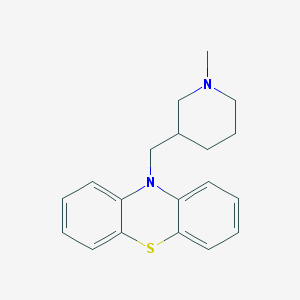 |
| Lipinski's druglikeness | This ligand has a partition coefficient log P greater than 5. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 137.0 nM | DrugMatrix in vitro pharmacology data | ChEMBL |
| Ki | 19.0 nM | DrugMatrix in vitro pharmacology data | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218