| Synonyms | NCGC00015273-04
HMS1920M03
Torazina
AX8120990
NCGC00015273-12
J10069
WLN: T C666 BN ISJ B3N1&1 EG
Bio1_001435
NCGC00024409-05
KBio2_005758
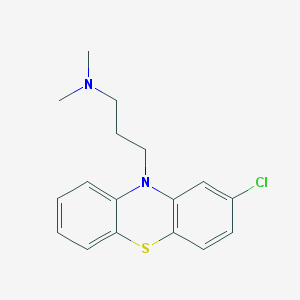
[3-(2-Chloro-phenothiazin-10-yl)-propyl]-dimethyl-amine( Chlorpromazine)
C17H19ClN2S
NSC 167745
KS-00002WW5
Chlor-Promanyl
Phenactyl
Chlorpromanyl (discontinued)
Prestwick0_000064
2-chloro-10-(3-(dimethylamino)propyl)-phenothiazine
Chlorpromazine;[3-(2-Chloro-phenothiazin-10-yl)-propyl]-dimethyl-amine
Propaphen
2-Cloro-10 (3-dimetilaminopropil)fenotiazina [Italian]
CPZ
Sanopron
34468-21-8
Largactilothiazine
DSSTox_CID_2808
SPBio_001111
AB00051943-15
MCULE-5976054678
Fenactil
SR-01000000012-5
Aminasine
Thorazine
Ampliactil
NCGC00015273-07
HMS3430C19
Tranzine (Salt/Mix)
BC 135
NCGC00015273-15
KBio1_000624
ZPEIMTDSQAKGNT-UHFFFAOYSA-N
BPBio1_001181
NCGC00024409-08
KBio3_002792
CCG-40059
NSC167745
KUC112482N
Chlorderazin
Phenothiazine, 2-chloro-10-(3-(dimethylamino)propyl)-
Chlorpromazine (USP/INN)
Prestwick3_000064
2-chloro-10-[3-(dimethylamino)propyl]-
Clorpromazina
Psychozine
3-(2-Chloro-10H-phenothiazin-10-yl)-N,N-dimethyl-1-propanamine
L000182
D00270
SCHEMBL8321
4560 Rp hydrochloride
Lopac-C-8138
DTXSID0022808
Spectrum2_001156
AB00051943_18
MolPort-001-727-953
FT-0653683
Hibernal (Salt/Mix)
Thorazine Spansule
AN-41563
NCGC00015273-10
I14-6226
Unitensen
Bio1_000457
NCGC00015273-19
KBio2_003190
[3-(2-Chloro-phenothiazin-10-yl)-propyl]-dimethyl-amine
BSPBio_002011
Novo-Chlorpromazine
KBioSS_000622
Oprea1_110255
Chloropromazine
Plegomazin
10H-Phenothiazine-10-propanamine, 2-chloro-N,N-dimethyl-, radical ion(1+)
Chlorpromazine [USAN:INN:BAN]
Promapar
2-Chloropromazine
cMAP_000017
R1242
3-(2-chlorophenothiazin-10-yl)-N,N-dimethyl-propan-1-amine
Largactil Liquid
DB00477
SKF-2601
M-1209
Epitope ID:136898
Spectrum5_000717
AKOS001490972
NCGC00015273-02
NCGC00015273-05
HMS2089C12
Tox21_110120
BAS 00010434
NCGC00015273-13
JHICC02042
Z80
Biomol-NT_000020
NCGC00024409-06
KBio2_007448
[3-(2-Chloro-phenothiazin-10-yl)-propyl]-dimethyl-amine(clorpromazine)
CAS-50-53-3
NSC-167745
KS-5101
Chlor-PZ
Phenathyl
(chloropromazine) [3-(2-Chloro-phenothiazin-10-yl)-propyl]-dimethyl-amine
Chlorpromazin
Prestwick1_000064
2-Chloro-10-(3-(dimethylamino)propyl)phenothiazine
Chlorpromazinum
Propaphenin
2601-A
Cromedazine
SBI-0050237.P004
4560 R.P
Largactyl
DSSTox_GSID_22808
SPBio_002168
AB00051943-16
Megaphen
Fenaktyl
SR-01000000012-6
Aminazin
Thorazine (TN)
Amplicitil
NCGC00015273-08
HMS501P06
U42B7VYA4P
BCP03610
NCGC00015273-16
KBio2_000622
[3-(2-chloro-10H-phenothiazin-10-yl)propyl]dimethylamine
BRD-K89997465-001-05-3
NINDS_000624
KBioGR_000806
CCRIS 3711
NSC17479
Chloro-3 (dimethylamino-3 propyl)-10 phenothiazine
Phenothiazine, 2-chloro-10-[3-(dimethylamino)propyl]-
10-(3-Dimethylaminopropyl)-2-chlorophenothiazine
Chlorpromazine cation radical
Proma
2-Chloro-10-[3-(dimethylamino)propyl]phenothiazine
Clorpromazina [INN-Spanish]
QSPL 401
3-(2-Chloro-10H-phenothiazin-10-yl)-N,N-dimethyl-1-propanamine #
Largactil
D01ZII
SKF 2601-A
50-53-3
Lopac0_000249
EINECS 200-045-8
Spectrum3_000346
AB0015094
N-(3-Dimethylaminopropyl)-3-chlorophenothiazine
GG 407
HL 5746
Thorazine Suppositories
API0001970
NCGC00015273-11
IDI1_000624
Wintermin
Bio1_000946
NCGC00024409-04
KBio2_004880
[3-(2-Chloro-phenothiazin-10-yl)-propyl]-dimethyl-amine (chlor-promazine)
C06906
Novomazina
KBioSS_002314
CHEMBL71
Pharmakon1600-01500184
Chlorpromados
Prazilpromactil
1N,1N-dimethyl-3-(2-chloro-10H-10-phenothiazinyl)-1-propanamine
Chlorpromazine [USP:INN:BAN]
Promazil
2-Cloro-10 (3-dimetilaminopropil)fenotiazina
Contomin
RP-4560
3-(2-chlorophenothiazin-10-yl)-N,N-dimethylpropan-1-amine
Largactil Oral Drops
DivK1c_000624
SMR001453710
AB00051943
M176
Esmind
Spectrum_000142
ALBB-022464
NCGC00015273-03
Aminazine
NCGC00015273-06
HMS2091E06
Tox21_110120_1
BBL028251
NCGC00015273-14
KB-232587
ZINC44027
BPBio1_000273
NCGC00024409-07
KBio3_001231
CAS-69-09-0
NSC-756689
KSC-315-032-
Chlordelazine
Phenothiazine hydrochloride
(chlorpromazine)[3-(2-Chloro-phenothiazin-10-yl)-propyl]-dimethyl-amine
Prestwick2_000064
2-Chloro-10-(3-dimethylaminopropyl)phenothiazine
Chlorpromazinum [INN-Latin]
Prozil
2601A
CTK1C5154
SC-50091
4560 R.P.
Lomazine (Salt/Mix)
DSSTox_RID_76736
SPECTRUM1500184
AB00051943_17
MLS003166901
Fraction AB
STK182870
Thorazine hydrochloride
Amplictil
NCGC00015273-09
HSDB 3033
UNII-U42B7VYA4P
BDBM50001888
NCGC00015273-17
KBio2_002312
[3-(2-chloro-10H-phenothiazin-10-yl)propyl]dimethylamine hydrochloride
BSPBio_000247
Norcozine (Salt/Mix)
KBioGR_002312
CHEBI:3647
NSC756689
Chloro-3 (dimethylamino-3 propyl)-10 phenothiazine [French]
Plegomasine
10H-Phenothiazine-10-propanamine, 2-chloro-N,N-dimethyl-
Chlorpromazine Tannate
Promactil
2-chloro-N,N-dimethyl-10H-Phenothiazine-10-propanamine
Clorpromazina [Italian]
QTL1_000021
3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine
Largactil (TN)
SKF 2601A
LS-105361
Elmarin
Spectrum4_000283
AC1L1EBT
NCGC00015273-01
GTPL83 [ Show all ] |
|---|


![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218