| Synonyms | IDI1_002631
Acetamide, N-[2-(5-methoxyindol-3-yl)ethyl]- (6CI,8CI)
KBio2_005801
ANW-42685
KSC498C4R
BRD-K97530723-001-07-6
M-1250
CCRIS 3472
Melatonex, Melatonin
CTK3J8148
Melatonin, >=99.5%
DivK1c_000353
F1929-1777
5-22-12-00042 (Beilstein Handbook Reference)
HMS1569K18
AB00053279_12
HMS3713K18
Acetamide, N-(2-(5-methoxy-1H-indol-3-yl)ethyl)-
NINDS_000353
Oprea1_814234
PREVENTION 1 (MELATONIN) (PREVENTION 1)
Melatonina (TN)
RTC-063565
MLS001055382
Sleep Right
N-acetyl-5-methoxy-tryptamine Melatonine
Spectrum4_000066
Nature'S Harmony
ST057260
NCGC00015680-08
Tox21_201527
NCGC00015680-16
[3H]-melatonin
NCGC00090727-07
JL5DK93RCL
Acetamide, {N-[2-(5-methoxyindol-3-yl)ethyl]-}
KBioSS_000665
BC200187
Lopac0_000787
BSPBio_000536
Mela-T
CHEMBL45
Melatonin (MLT)
D0B5CB
DSSTox_GSID_22421
GTPL1357
8041-44-9
HMS2096K18
AC-10019
HSDB 7509
Acetamide, N-(2-(5-methoxyindol-3-yl)ethyl)-
NCGC00256404-01
NSC-113928
Prestwick0_000458
PREVENTION 4 (MELATONIN)
Melatonine;
SBB003265
MT6
SPBio_001527
N-[2-(5-Methoxy-1H-indol-3-yl)ethyl)acetamide
SR-01000075559
NCGC00015680-03
SW196607-4
NCGC00015680-11
UNII-JL5DK93RCL
NCGC00090727-02
{N-[2-(5-methoxy-1H-indol-3-yl)ethyl]-} Acetamide
Acetamide, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]-(9CI)
KBio2_000665
AK110931
KS-000048J8
BIDD:ER0618
M 5250
CAS-73-31-4
Melatonex
CM-539
Melatonin 1.0 mg/ml in Methanol
DB01065
EINECS 200-797-7
4-ACETAMIDO-4'-ISOTHIO-CYANATOSTILBENE-2,2'-DISULFONIC ACID
Guna-dermo (Salt/Mix)
AB00053279
HMS3370J20
AC1Q4F1X
I10-0345
NCI60_004378
NSC56423
Prestwick3_000458
Melatonin, United States Pharmacopeia (USP) Reference Standard
Revital Melatonin
ML1
SDCCGMLS-0065812.P001
N-(2-(5-Methoxyindol-3-yl)ethyl)acetamide
Spectrum2_001344
N-[2-(5-methoxyindol-3-yl)ethyl]-Acetamide
SR-01000075559-7
NCGC00015680-06
Tox21_110195
NCGC00015680-14
ZB001948
NCGC00090727-05
IN1244
Acetamide, N-[2-(5-methoxyindol-3-yl)ethyl]-(6CI,8CI)
KBio3_002226
ARONIS24462
L001261
BRD-K97530723-001-11-8
M1105
CHEBI:16796
D08170
Melatonin, British Pharmacopoeia (BP) Reference Standard
DRLFMBDRBRZALE-UHFFFAOYSA-N
005M655
FT-0628191
5-Methoxy-N-acetyltryptamine
HMS1921E04
AB0007332
HMS501B15
Acetamide, N-(2-(5-methoxy-1H-indol-3-yl)ethyl)- (9CI)
NMR/14327425
Pineal Hormone
PREVENTION 2 (MELATONIN)
Melatonine
Rx Balance
MLS001240204
SMP2_000309
N-Acetyl-5-methoxytryptamine
Spectrum5_001745
NCGC00015680-01
ST24031293
NCGC00015680-09
Tox21_302926
NCGC00015680-18
[3H]melatonin
NCGC00090727-08
Acetamide, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]-
KB-178582
ACT03490
KM1545
BCP28154
LP00787
BSPBio_003006
Melapure
ChemDiv2_003916
Melatonin (synth.) standard-grade
D0M9SI
DSSTox_RID_76585
GTPL224
HMS2233D23
AC1L1A9Q
HY-B0075
NCGC00259077-01
NSC-56423
Prestwick1_000458
PREVENTION 5 (MELATONIN)
Melovine
SC-16475
N-(2-(5-Methoxy-1H-indol-3-yl)ethyl)acetamide
SPBio_002475
N-[2-(5-methoxy-1H-indol-3-yl)ethyl]-Acetamide
SR-01000075559-1
NCGC00015680-04
TC-063565
NCGC00015680-12
Vivitas
NCGC00090727-03
{N-[2-(5-methoxyindol-3-yl)ethyl]-} Acetamide
Acetamide, N-[2-(5-methoxyindol-3-yl)ethyl]-
KBio2_003233
AKOS000276269
KS-1454
BPBio1_000590
M-1200
CCG-38837
Melatonex, 73-31-4
CS-1769
Melatonin solution, 1.0 mg/mL in methanol, ampule of 1 mL, certified reference material
Disodium 4-acetamido-4'-isothiocyanato-stilben-2,2'-disulfonate
EU-0100787
4CN-3100
HMS1380B22
AB00053279-10
HMS3654A22
ACE073
IDI1_000353
Night Rest
Oprea1_104553
Prestwick_312
Melatonina
RP17870
MLS000859594
SDCCGMLS-0065812.P002
N-acetyl-5-methoxy-tryptamine
Spectrum3_001393
N-[2-(5-methoxyindol-3-yl)ethyl]acetamide
SR-01000075559-8
NCGC00015680-07
Tox21_110195_1
NCGC00015680-15
ZINC57060
NCGC00090727-06
J10164
Acetamide, {N-[2-(5-methoxy-1H-indol-3-yl)ethyl]-}
KBioGR_000591
BAS 01281092
Lopac-M-5250
BRN 0205542
MCULE-1497884152
Melatonin (JAN)
D0AN7B
Melatonin, Pharmaceutical Secondary Standard; Certified Reference Material
DSSTox_CID_2421
0E2B08C1-B325-45B1-8939-6F9081EFDFA4
FT-0658928
73-31-4
HMS2089F09
AB2000011
HSCI1_000400
Acetamide, N-(2-(5-methoxy-1H-indol-3-yl)ethyl)-(9CI)
NSC 113928
Posidorm
PREVENTION 3 (MELATONIN)
Melatonine, 99%
S1204
MolPort-000-737-883
SMR000326666
N-[2-(5-Methoxy-1H-indol-3-yl)-ethyl]-acetamide
Spectrum_000185
NCGC00015680-02
STK386880
NCGC00015680-10
Tox21_500787
NCGC00090727-01
[3H]MLT
NCGC00090727-09
Acetamide, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]- (9CI)
KBio1_000353
AJ-09558
KS-00000GX9
BDBM9019
LS-1623
C01598
Melatol
Circadin
Melatonin (synth.) ultra-pure
DTXSID1022421
3-(n-acetyl-2-aminoethyl)-5-methoxyindole
Guna-dermo
A4039/0172195
HMS3262M16
AC1Q4F1W
I05-0076
NCGC00261472-01
NSC113928
Prestwick2_000458
Melatonin, powder, >=98% (TLC)
PubChem18245
MFCD00005655
SCHEMBL19018
N-(2-(5-methoxyindol-3-yl)ethyl)-Acetamide
SPECTRUM1500690
N-[2-(5-Methoxy-1H-indol-3-yl)ethyl]acetamide
SR-01000075559-6
NCGC00015680-05
TNP00300
NCGC00015680-13
WLN: T56 BMJ D2MV1 GO1
NCGC00090727-04 [ Show all ] |
|---|
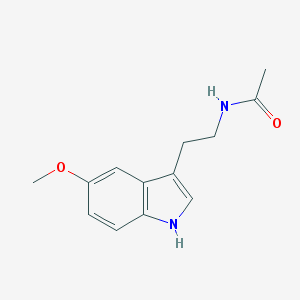
| Inchi ID | InChI=1S/C13H16N2O2/c1-9(16)14-6-5-10-8-15-13-4-3-11(17-2)7-12(10)13/h3-4,7-8,15H,5-6H2,1-2H3,(H,14,16) |
|---|


![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218