

You can:
| Name | D(2) dopamine receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | DRD2 |
| Synonym | dopamine receptor 2 Dopamine D2 receptor D2R D2A and D2B D2(415) and D2(444) [ Show all ] |
| Disease | Substance dependence Major depressive disorder Maintain blood pressure in hypotensive states Insomnia Inflammatory disease [ Show all ] |
| Length | 443 |
| Amino acid sequence | MDPLNLSWYDDDLERQNWSRPFNGSDGKADRPHYNYYATLLTLLIAVIVFGNVLVCMAVSREKALQTTTNYLIVSLAVADLLVATLVMPWVVYLEVVGEWKFSRIHCDIFVTLDVMMCTASILNLCAISIDRYTAVAMPMLYNTRYSSKRRVTVMISIVWVLSFTISCPLLFGLNNADQNECIIANPAFVVYSSIVSFYVPFIVTLLVYIKIYIVLRRRRKRVNTKRSSRAFRAHLRAPLKGNCTHPEDMKLCTVIMKSNGSFPVNRRRVEAARRAQELEMEMLSSTSPPERTRYSPIPPSHHQLTLPDPSHHGLHSTPDSPAKPEKNGHAKDHPKIAKIFEIQTMPNGKTRTSLKTMSRRKLSQQKEKKATQMLAIVLGVFIICWLPFFITHILNIHCDCNIPPVLYSAFTWLGYVNSAVNPIIYTTFNIEFRKAFLKILHC |
| UniProt | P14416 |
| Protein Data Bank | 6cm4, 6c38 |
| GPCR-HGmod model | P14416 |
| 3D structure model | This structure is from PDB ID 6cm4. |
| BioLiP | BL0408379, BL0403379 |
| Therapeutic Target Database | T67162 |
| ChEMBL | CHEMBL217 |
| IUPHAR | 215 |
| DrugBank | BE0000756 |
| Name | pramipexole |
|---|---|
| Molecular formula | C10H17N3S |
| IUPAC name | (6S)-6-N-propyl-4,5,6,7-tetrahydro-1,3-benzothiazole-2,6-diamine |
| Molecular weight | 211.327 |
| Hydrogen bond acceptor | 4 |
| Hydrogen bond donor | 2 |
| XlogP | 1.9 |
| Synonyms | FT-0651622 K-0425 Mirapex (6S)-N6-propyl-4,5,6,7-tetrahydro-1,3-benzothiazole-2,6-diamine NCGC00167441-01 [ Show all ] |
| Inchi Key | FASDKYOPVNHBLU-ZETCQYMHSA-N |
| Inchi ID | InChI=1S/C10H17N3S/c1-2-5-12-7-3-4-8-9(6-7)14-10(11)13-8/h7,12H,2-6H2,1H3,(H2,11,13)/t7-/m0/s1 |
| PubChem CID | 119570 |
| ChEMBL | CHEMBL301265 |
| IUPHAR | 953 |
| BindingDB | 50116766 |
| DrugBank | DB00413 |
Structure | 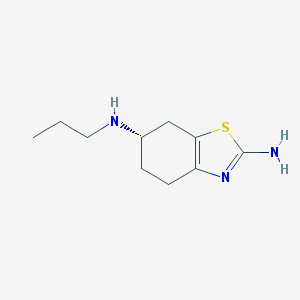 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| Activity | <75.0 % | None | ChEMBL |
| Activity | 95.0 % | PMID23237836 | ChEMBL |
| EC50 | 5.0 nM | PMID18313303 | BindingDB,ChEMBL |
| EC50 | 7.079 nM | None | ChEMBL |
| EC50 | 27.0 nM | PMID17976986 | ChEMBL |
| IC50 | <10000.0 nM | PMID23237836 | BindingDB,ChEMBL |
| Intrinsic activity | 100.0 % | PMID18313303 | ChEMBL |
| Ki | 21.0 nM | PMID15801839, PMID16134944 | BindingDB,ChEMBL |
| Ki | 27.0 nM | PMID12161137 | BindingDB,ChEMBL |
| Ki | 39.8107 - 7943.28 nM | PMID7756621, PMID7664822 | IUPHAR |
| Ki | 40.0 nM | PMID12161137 | BindingDB,ChEMBL |
| Ki | 139.0 nM | PMID11000009 | ChEMBL |
| Ki | 1900.0 nM | PMID15801839, PMID16134944 | BindingDB,ChEMBL |
| Ki | 6300.0 nM | PMID15801839, PMID16134944 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218