

You can:
| Name | G-protein coupled receptor 12 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | GPR12 |
| Synonym | GPR12 GPCR21 Gpcr20 GPCR12 Gpcr01 [ Show all ] |
| Disease | N/A |
| Length | 334 |
| Amino acid sequence | MNEDLKVNLSGLPRDYLDAAAAENISAAVSSRVPAVEPEPELVVNPWDIVLCTSGTLISCENAIVVLIIFHNPSLRAPMFLLIGSLALADLLAGIGLITNFVFAYLLQSEATKLVTIGLIVASFSASVCSLLAITVDRYLSLYYALTYHSERTVTFTYVMLVMLWGTSICLGLLPVMGWNCLRDESTCSVVRPLTKNNAAILSVSFLFMFALMLQLYIQICKIVMRHAHQIALQHHFLATSHYVTTRKGVSTLAIILGTFAACWMPFTLYSLIADYTYPSIYTYATLLPATYNSIINPVIYAFRNQEIQKALCLICCGCIPSSLAQRARSPSDV |
| UniProt | P47775 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P47775 |
| 3D structure model | This predicted structure model is from GPCR-EXP P47775. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | N/A |
| IUPHAR | 86 |
| DrugBank | BE0009153 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 553612 | 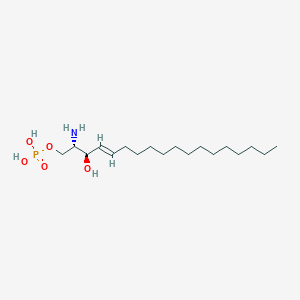 Sphingosine 1-phosphate Sphingosine 1-phosphate | C18H38NO5P | 379.478 | 6 / 4 | 1.9 | Yes |
| 554037 | 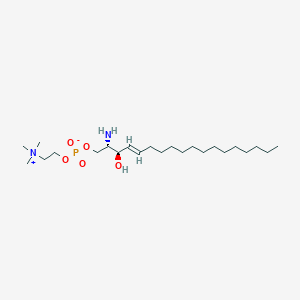 sphingosylphosphorylcholine sphingosylphosphorylcholine | C23H49N2O5P | 464.628 | 6 / 2 | 4.7 | Yes |
| 551352 | 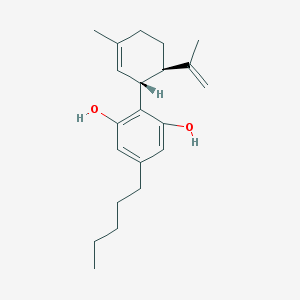 cannabidiol cannabidiol | C21H30O2 | 314.469 | 2 / 2 | 6.5 | No |
| 551911 | 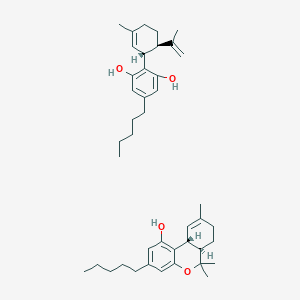 Cannabinoids Cannabinoids | C42H60O4 | 628.938 | 4 / 3 | N/A | No |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218