

You can:
| Name | Leukotriene B4 receptor 1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | LTB4R |
| Synonym | BLT1 receptor BLTR Chemoattractant receptor-like 1 G-protein coupled receptor 16 GPR16 [ Show all ] |
| Disease | Inflammatory disease Inflammatory bowel disease Human immunodeficiency virus infection Pancreatic cancer Psoriasis [ Show all ] |
| Length | 352 |
| Amino acid sequence | MNTTSSAAPPSLGVEFISLLAIILLSVALAVGLPGNSFVVWSILKRMQKRSVTALMVLNLALADLAVLLTAPFFLHFLAQGTWSFGLAGCRLCHYVCGVSMYASVLLITAMSLDRSLAVARPFVSQKLRTKAMARRVLAGIWVLSFLLATPVLAYRTVVPWKTNMSLCFPRYPSEGHRAFHLIFEAVTGFLLPFLAVVASYSDIGRRLQARRFRRSRRTGRLVVLIILTFAAFWLPYHVVNLAEAGRALAGQAAGLGLVGKRLSLARNVLIALAFLSSSVNPVLYACAGGGLLRSAGVGFVAKLLEGTGSEASSTRRGGSLGQTARSGPAALEPGPSESLTASSPLKLNELN |
| UniProt | Q15722 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q15722 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q15722. |
| BioLiP | N/A |
| Therapeutic Target Database | T59626 |
| ChEMBL | CHEMBL3911 |
| IUPHAR | 267 |
| DrugBank | BE0003490 |
| Name | LY223982 |
|---|---|
| Molecular formula | C30H30O7 |
| IUPAC name | 3-[3-(2-carboxyethyl)-4-[(E)-6-(4-methoxyphenyl)hex-5-enoxy]benzoyl]benzoic acid |
| Molecular weight | 502.563 |
| Hydrogen bond acceptor | 7 |
| Hydrogen bond donor | 2 |
| XlogP | 5.8 |
| Synonyms | 3-{[3-(2-carboxyethyl)-4-{[(5E)-6-(4-methoxyphenyl)hex-5-en-1-yl]oxy}phenyl]carbonyl}benzoic acid Cgs 23131 HY-112737 LY-223980 SCHEMBL7202040 [ Show all ] |
| Inchi Key | SYZSSLLFRVDRHL-QPJJXVBHSA-N |
| Inchi ID | InChI=1S/C30H30O7/c1-36-26-14-10-21(11-15-26)7-4-2-3-5-18-37-27-16-12-24(19-22(27)13-17-28(31)32)29(33)23-8-6-9-25(20-23)30(34)35/h4,6-12,14-16,19-20H,2-3,5,13,17-18H2,1H3,(H,31,32)(H,34,35)/b7-4+ |
| PubChem CID | 6444688 |
| ChEMBL | CHEMBL49302 |
| IUPHAR | 3415 |
| BindingDB | 50001611 |
| DrugBank | N/A |
Structure | 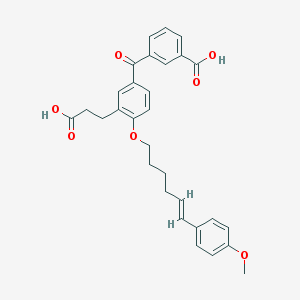 |
| Lipinski's druglikeness | This ligand is heavier than 500 daltons. This ligand has a partition coefficient log P greater than 5. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 1.9 nM | PMID8389876 | BindingDB,ChEMBL |
| IC50 | 2.0 nM | PMID1333012, PMID1333011 | BindingDB,ChEMBL |
| IC50 | 3.5 nM | PMID1333012, PMID1333011 | BindingDB,ChEMBL |
| IC50 | 12.0 nM | PMID8410983, PMID1331462, PMID7932560 | BindingDB,ChEMBL |
| IC50 | 13.0 nM | , Bioorg. Med. Chem. Lett., (1993) 3:10:1981, PMID8389876, PMID8709092 | BindingDB,ChEMBL |
| IC50 | 13.2 nM | PMID8389876, PMID2170648 | BindingDB,ChEMBL |
| IC50 | 1000.0 nM | PMID1333012 | BindingDB,ChEMBL |
| Kd | 13.0 nM | PMID1320692 | IUPHAR |
| Ki | 7.8 nM | , Bioorg. Med. Chem. Lett., (1993) 3:10:1981 | BindingDB,ChEMBL |
| Ki | 360.0 nM | PMID8230123 | BindingDB,ChEMBL |
| Relative potency | 159.0 - | PMID8389876 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218