


GPCR-RD is a database for experimental restaints of GPCRs. Although no solved X-ray or NMR structure is available for most GPCR sequences, numerous experiments have been performed on GPCRs to identify the critical residues and motifs. Experimental restraints were systematically collected from the literature using an automated text mining algorithm combined with manual validation, with the purpose of assisting GPCR 3-D structure modeling and function annotation. The GPCR-RD data are useful for GPCR 3D structure modeling, such as the GPCR-I-TASSER algorithm. The flow chart below summarizes the procedure for the extraction of experimental data and spatial restraints from literature and other resources. 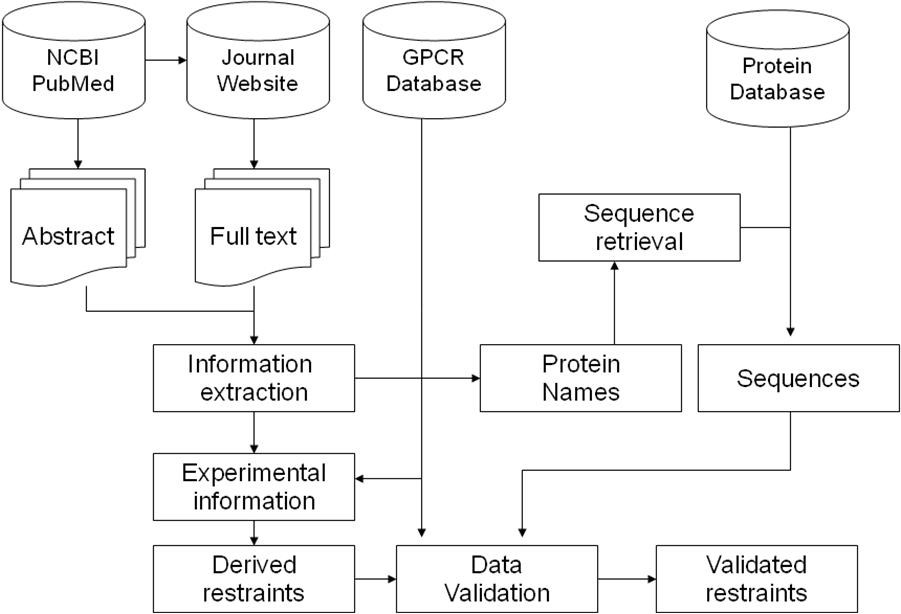
|
Other GPCR-related resources
|
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218